

| What is the COTH server? |
COTH server is an internet service for prediction of protein complex structure prediction using a threading based approach. While most efforts in the field of protein complex structure prediction is concentrated on protein-protein docking, it suffers from a severely limited in scope since docking can only be performed when the unbound protein structures are known before hand. COTH aims to bridge this gap by predicting protein complex structures from the primary amino acid sequence by the aid of identification of protein complex template structures.
| How does COTH identify the best templates? |
When the users submit a protein complex sequence, the COTH server attempts to identify a complex sequence from a protein complex structure library which is most homologous/ structually analogous to the submitted query sequence. COTH attempts to identify the best template using a profile-profile alignment based approach where the scoring function includes a number of predicted structural features to aid profile-profile alignment. The scoring functions include 1) Match between the sequence profiles of the query and the template 2) Match between the predicted secondary structure of the query and the native secondary structure of the template. 3) Match between the sequence profile of the query and a depth based structure profile of the templte. 3) Match between the predicted solvent accessibility of the query residues and the native solvent accessibility of the template residues. 4) Match between the predicted torsional angles (phi and psi angles) of the query residues and the native torsional angles of the templat e residues. 5)Match between the hydrophcity of the query and template residues 6) Match between the predicted interface residues of the query and the native interface residues of the template complex. In order to prevent cross-alignment of chains,i.e. both chains of one complex aligning to a single chain of the other complex we use a modified dynamic programing approach (See Figure 1) in order to prevent cross-alignment.
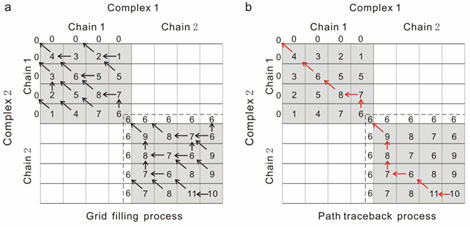
After the query sequence is aligned with all complexes in the protein complex library the templates are
ranked based on mean Zscore given by:
| How does COTH remedy the incompleteness of the protein complex structure library? |
The protein complex structure library is indeed incomplete. To remedy this problem, the COTH server simultaneously runs the MUSTER single chain threading for both the chains of the query complex. Since MUSTER is run on the much larger protein tertiary structure library the templates identified by MUSTER for the individual chains are of better quality than that identified by COTH threading only. Following the generation of the MUSTER individual chain templates, therefore the COTH server superimposes the MUSTER templates onto the corresponding chain of the multimeric template framework. This ensures a much improved quality of the individual chains while maintaining the orientation information identified by multimeric threading. To understand the complete flow of the COTH algorithm refer to Figure 2.
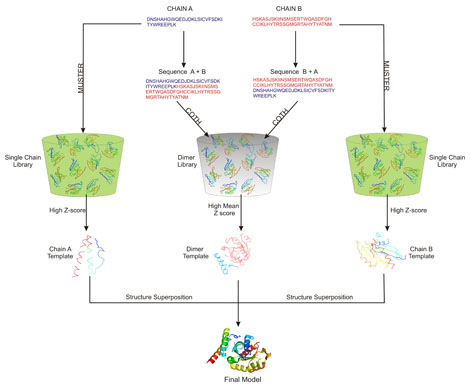
| How to cite COTH |
You are requested to cite following articles when you use the I-TASSER server:
| Contact information |
The COTH server is in active development with the goal to provide the reliable protein complex structure predictions. Please help us achieve the goal by sending your questions, feedback, and comments to: yangzhanglab@umich.edu.
yangzhanglab![]() umich.edu
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
umich.edu
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218