

You can:
| Name | Neurotensin receptor type 1 |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | NTSR1 |
| Synonym | NTSR1 NTS1 receptor NTRH NTR1 NTR [ Show all ] |
| Disease | Acute or chronic pain Alcohol use disorders Pain Inflammatory bowel disease Neurological disease |
| Length | 418 |
| Amino acid sequence | MRLNSSAPGTPGTPAADPFQRAQAGLEEALLAPGFGNASGNASERVLAAPSSELDVNTDIYSKVLVTAVYLALFVVGTVGNTVTAFTLARKKSLQSLQSTVHYHLGSLALSDLLTLLLAMPVELYNFIWVHHPWAFGDAGCRGYYFLRDACTYATALNVASLSVERYLAICHPFKAKTLMSRSRTKKFISAIWLASALLAVPMLFTMGEQNRSADGQHAGGLVCTPTIHTATVKVVIQVNTFMSFIFPMVVISVLNTIIANKLTVMVRQAAEQGQVCTVGGEHSTFSMAIEPGRVQALRHGVRVLRAVVIAFVVCWLPYHVRRLMFCYISDEQWTPFLYDFYHYFYMVTNALFYVSSTINPILYNLVSANFRHIFLATLACLCPVWRRRRKRPAFSRKADSVSSNHTLSSNATRETLY |
| UniProt | P30989 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | P30989 |
| 3D structure model | This predicted structure model is from GPCR-EXP P30989. |
| BioLiP | N/A |
| Therapeutic Target Database | T02728 |
| ChEMBL | CHEMBL4123 |
| IUPHAR | 309 |
| DrugBank | N/A |
| Name | MLS000663547 |
|---|---|
| Molecular formula | C17H16N6O5 |
| IUPAC name | 1-(4-morpholin-4-yl-2,1,3-benzoxadiazol-7-yl)-3-(4-nitrophenyl)urea |
| Molecular weight | 384.352 |
| Hydrogen bond acceptor | 8 |
| Hydrogen bond donor | 2 |
| XlogP | 1.3 |
| Synonyms | HMS2697L23 SR-01000276120-1 BDBM50480 MolPort-001-589-859 ZINC13153222 [ Show all ] |
| Inchi Key | BJJUETIRJVEVLQ-UHFFFAOYSA-N |
| Inchi ID | InChI=1S/C17H16N6O5/c24-17(18-11-1-3-12(4-2-11)23(25)26)19-13-5-6-14(16-15(13)20-28-21-16)22-7-9-27-10-8-22/h1-6H,7-10H2,(H2,18,19,24) |
| PubChem CID | 2955071 |
| ChEMBL | CHEMBL1482308 |
| IUPHAR | N/A |
| BindingDB | 50480 |
| DrugBank | N/A |
Structure | 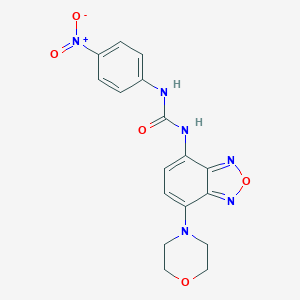 |
| Lipinski's druglikeness | This ligand satisfies Lipinski's rule of five. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| EC50 | <40000.0 nM | , PubChem BioAssay data set | BindingDB,ChEMBL |
zhanglab![]() zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218