| Synonyms | CM0152
Mirtasole (TN)
EN300-49851
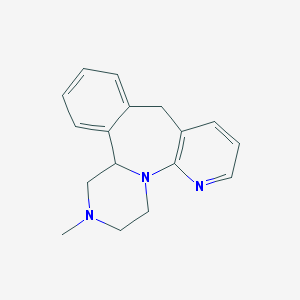
1,2,3,4,10,14b-Hexahydro-2-methylpyrazino[2,1-a]pyrido[2,3-c][2]benzazepine
GTPL7241
5-methyl-2,5,19-triazatetracyclo[13.4.0.0?,?.0?,??]nonadeca-1(19),8,10,12,15,17-hexaene
HMS3657M13
85650-52-8
KS-1086
AKOS005530681
ME-2040
Arintapina (TN)
Miralix (TN)
BCP22244
Mirtachem (TN)
CAS-61337-67-5
Mirtamerck (TN)
Mirtazapine (JAN/USAN/INN)
SMR000466347
Mirtazapine, 98%
Tarzapine (TN)
Mirtazep (TN)
Valdren (TN)
Mirtoral (TN)
Zispin SolTab (TN)
MLS000759460
NCGC00025346-02
PDSP1_001529
Pyrazino(2,1-a)pyrido(2,3-c)(2)benzazepine, 1,2,3,4,10,14b-hexahydro-2-methyl-
Remergon (TN)
Mirtaz
Rexer
Mirtapin(TN)
CPD000466347
Mirtatifi (TN)
DSSTox_CID_3325
(14bR)-2-methyl-1,2,3,4,10,14b-hexahydropyrazino[2,1-a]pyrido[2,3-c][2]benzazepine
Finpharma (TN)
2-Methyl-1,2,3,4,10,14b-hexahydropyrazino[2,1-a]pyrido[2,3-c][2]benzazepine
HMS2233K03
I01-1882
AB2000422
LS-127703
Amirel (TN)
Mepirzapine
Axit
Miro (TN)
Bexzis (TN)
Mirtagamma (TN)
Celltech brand of mirtazapine
Mirtazapine for system suitability, European Pharmacopoeia (EP) Reference Standard
SR-01000597530-1
Mirtazapine, United States Pharmacopeia (USP) Reference Standard
Tazascope (TN)
Mirtazipine
Zapex (TN)
Mirzaten (TN)
MLS006011449
Norset (TN)
Pharmazepine (TN)
Pyrazino[2,1-a]pyrido[2,3-c][2]benzazepine, 1,2,3,4,10,14b-hexahydro-2-methyl-, (.+/-.)-
Remeron SolTab
Mirtazapin (TN)
s2016
CHEMBL654
Mirtaron (TN)
D00563
DTXSID0023325
1,2,3,4,10,14b-Hexahydro-2-methylpyrazino(2,1-a)pyrido(2,3-c)benzazepine
FT-0628951
5-methyl-2,5,19-triazatetracyclo[13.4.0.0(2),.0,(1)(3)]nonadeca-1(19),8,10,12,15,17-hexaene
HMS3374J01
K532
AC1Q3ZYM
M2151
ANW-42478
MFCD00865427
BBL009962
Mirtabene
C07570
Mirtalphagen (TN)
SCHEMBL35408
Mirtazapine [USAN:INN:BAN]
STK711107
Mirtazelon (TN)
TR-026749
Mirtazza (TN)
Zispin
Mizapin (TN)
NC00404
Org-3770
PubChem22104
Remergil (TN)
Mirtatsapiini (TN)
Remeron;Avanza
Mirtazapina [INN-Spanish]
Mirtapax (TN)
Combar (TN)
Mirtastad (TN)
DB00370
(+/-)-12-Methyl-1,2,3,4,9,13b-hexahydro-2,4a,5-triaza-tribenzo[a,c,e]cycloheptene
Esprital (TN)
1,2,3,4,10,14b-Hexahydro-2-methylpyrazino[2,1-a]pyrido[2,3-c][2]benzazepine.
Hexazipin (TN)
5-methyl-2,5,19-triazatetracyclo[13.4.0.0^{2,7}.0^{8,13}]nonadeca-1(15),8(13),9,11,16,18-hexaene
HMS3713P13
AB0014270
L001294
Alphamirt (TN)
Medizapin (TN)
Avanza
Mirap (TN)
BCP9000930
Mirtacur (TN)
CCG-101154
Mirtazapine (Remeron, Avanza)
SolTab (TN)
Mirtazapine, >=98% (HPLC)
Tazamel (TN)
Mirtazepin (TN)
Vastat (TN)
Mirzagen (TN)
Zistap. (TN)
MLS001076676
NE42580
PDSP2_001513
Pyrazino[2,1-a]pyrido[2,3-c][2]benzazepine, 1,2,3,4,10,14b-hexahydro-2-methyl-
Remeron
Mirtaz (TN)
Rexer (TN)
Mirtaratio (TN)
CS-2398
DSSTox_GSID_23325
(N-methyl-11C)mirtazapine
Finscope (TN)
2-methyl-1,2,3,4,9,13b-hexahydro-2,4a,5-triaza-tribenzo[a,c,e]cycloheptene
HMS3268F21
6-Azamianserin
I06-0606
AC-15480
LS-185067
AN-35645
Mepirzepine
Axit (TN)
Miron (TN)
BRD-A64977602-001-01-9
Mirtal (TN)
CHEBI:6950
SAM001246659
Mirtazapine solution, 1.0 mg/mL in methanol, ampule of 1 mL, certified reference material
SR-01000597530-4
Mirtazapinum
Tirzamed (TN)
Mirtazon
Zicomber (TN)
Mirzaten Q-Tab (TN)
MolPort-003-849-233
Noxibel (TN)
Promyrtil
Reflex (TN)
Remeron SolTab (TN)
Mirtazapina
Ciblex (TN)
Mirtascope (TN)
D05ZIK
EINECS 288-060-6
1,2,3,4,10,14b-Hexahydro-2-methylpyrazino[2,1-a]pyrido[2,3-c](2)benzazepine
Genamirt (TN)
5-methyl-2,5,19-triazatetracyclo[13.4.0.0;{2,7}.0;{8,13}]nonadeca-1(19),8(13),9,11,15,17-hexaene
HMS3394H03
82601-27-2
KS-000010W5
Afloyan (TN)
MCULE-1174883554
Arintapin (TN)
Mianserin, 6-Aza-; ORG 3770
BCP14560
Mirtabene (TN)
Calixta (TN)
Mirtamed (TN)
Smilon
Mirtazapine [USAN:USP:INN:BAN]
SW197784-4
Mirtazen (TN)
VA11306
Mirtel (TN)
Zispin (TN)
Mizapin Sol (TN)
NCGC00025346-01
Organon brand of mirtazapine
PYR432
Remergon
Mirtawin (TN)
Remirta (TN)
Mirtapharm (TN)
Comenter (TN)
Mirtastada (TN)
Divaril (TN)
(1)-1,2,3,4,10,14b-Hexahydro-2-methylpyrazino(2,1-a)pyrido(2,3-c)(2)benzazepine
Finmirtaza (TN)
2-methyl-1,2,3,4,10,14b-hexahydrobenzo[c]pyrazino[1,2-a]pyrido[3,2-f]azepine
HMS2052H03
5-methyl-2,5,19-triazatetracyclo[13.4.0.0^{2,7}.0^{8,13}]nonadeca-1(19),8(13),9,11,15,17-hexaene
HY-B0352
AB00698265_08
Loxozapin (TN)
Alphazagen (TN)
Mepirzapin
Avanza (TN)
Mirazep (TN)
BDBM50115644
Mirtadepi (TN)
CCG-220556
Mirtazapine 1.0 mg/ml in Methanol
SR-01000597530
Mirtazapine, European Pharmacopoeia (EP) Reference Standard
Tazapin (TN)
Mirtazepine
Z2327131613
Mirzalux (TN)
Zuleptan (TN)
MLS001424294
Norset
Pharmasole (TN)
Pyrazino[2,1-a]pyrido[2,3-c][2]benzazepine, 1,2,3,4,10,14b-hexahydro-2-methyl- 85650-52-8
Remeron (TN)
Mirtazapin
RONZAEMNMFQXRA-UHFFFAOYSA-N
Mirtaril (TN)
CTK8B3409
DSSTox_RID_76979
FT-0601544
337M675
HMS3370B05
61337-67-5
J10458
AC1L1HND
M-130
AN-8319
methylBLAH
Azamianserin
Mirta (TN)
BRD-A64977602-001-04-3
Mirtalich (TN)
SC-16155
Mirtazapine [USAN:BAN:INN]
ST24048108
Mirtazapinum [INN-Latin]
Tox21_110965
Mirtazon (TN)
Zismirt (TN)
Mitrazen (TN)
Mundogen brand of mirtazapine
ORG 3770
Promyrtil (TN)
Remergil
Remeron, Avanza, Zispin
Mirtazapina (TN) [ Show all ] |
|---|


![]() zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218