

You can:
| Name | D(1A) dopamine receptor |
|---|---|
| Species | Rattus norvegicus (Rat) |
| Gene | Drd1 |
| Synonym | D1 receptor D1A DADR dopamine D1 receptor Dopamine-1A receptor [ Show all ] |
| Disease | N/A for non-human GPCRs |
| Length | 446 |
| Amino acid sequence | MAPNTSTMDEAGLPAERDFSFRILTACFLSLLILSTLLGNTLVCAAVIRFRHLRSKVTNFFVISLAVSDLLVAVLVMPWKAVAEIAGFWPLGPFCNIWVAFDIMCSTASILNLCVISVDRYWAISSPFQYERKMTPKAAFILISVAWTLSVLISFIPVQLSWHKAKPTWPLDGNFTSLEDTEDDNCDTRLSRTYAISSSLISFYIPVAIMIVTYTSIYRIAQKQIRRISALERAAVHAKNCQTTAGNGNPVECAQSESSFKMSFKRETKVLKTLSVIMGVFVCCWLPFFISNCMVPFCGSEETQPFCIDSITFDVFVWFGWANSSLNPIIYAFNADFQKAFSTLLGCYRLCPTTNNAIETVSINNNGAVVFSSHHEPRGSISKDCNLVYLIPHAVGSSEDLKKEEAGGIAKPLEKLSPALSVILDYDTDVSLEKIQPVTHSGQHST |
| UniProt | P18901 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | N/A |
| 3D structure model | No available structures or models |
| BioLiP | N/A |
| Therapeutic Target Database | N/A |
| ChEMBL | CHEMBL265 |
| IUPHAR | 214 |
| DrugBank | N/A |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 85 | 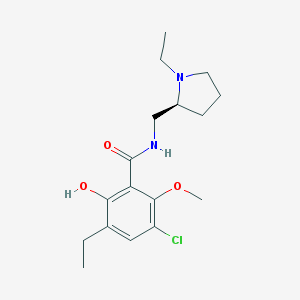 Eticlopride Eticlopride | C17H25ClN2O3 | 340.848 | 4 / 2 | 3.1 | Yes |
| 291 | 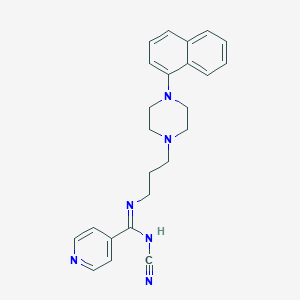 CHEMBL1783352 CHEMBL1783352 | C24H26N6 | 398.514 | 5 / 1 | 3.5 | Yes |
| 403 | 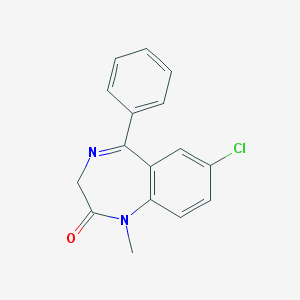 diazepam diazepam | C16H13ClN2O | 284.743 | 2 / 0 | 3.0 | Yes |
| 742 | 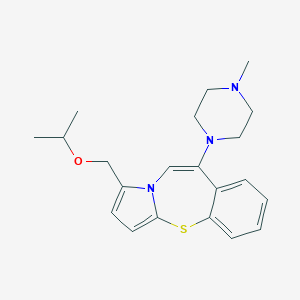 CHEMBL367875 CHEMBL367875 | C21H27N3OS | 369.527 | 4 / 0 | 3.4 | Yes |
| 1397 | 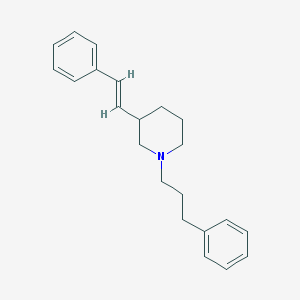 CHEMBL37661 CHEMBL37661 | C22H27N | 305.465 | 1 / 0 | 5.5 | No |
| 1399 | 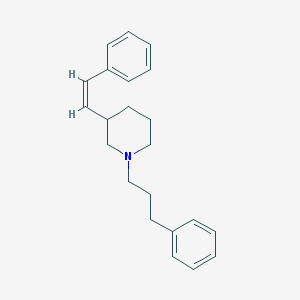 CHEMBL39483 CHEMBL39483 | C22H27N | 305.465 | 1 / 0 | 5.5 | No |
| 1645 | 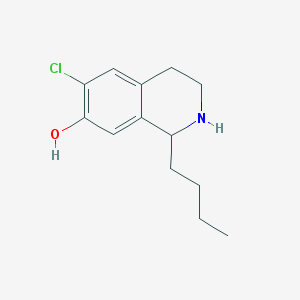 CHEMBL574111 CHEMBL574111 | C13H18ClNO | 239.743 | 2 / 2 | 3.4 | Yes |
| 1810 | 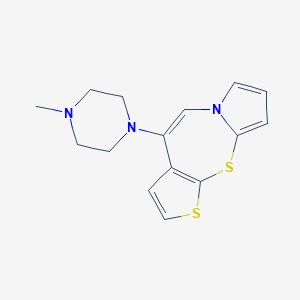 CHEMBL180319 CHEMBL180319 | C15H17N3S2 | 303.442 | 4 / 0 | 3.0 | Yes |
| 3191 | 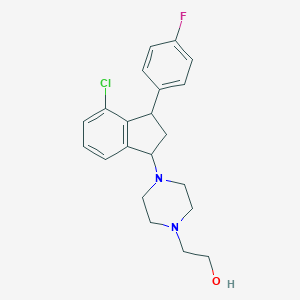 CHEMBL92896 CHEMBL92896 | C21H24ClFN2O | 374.884 | 4 / 1 | 3.5 | Yes |
| 3417 | 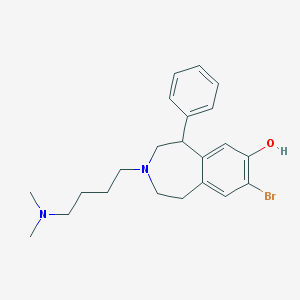 CHEMBL135616 CHEMBL135616 | C22H29BrN2O | 417.391 | 3 / 1 | 4.8 | Yes |
| 3753 | 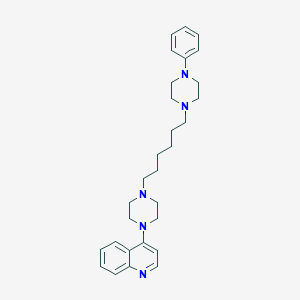 CHEMBL1173017 CHEMBL1173017 | C29H39N5 | 457.666 | 5 / 0 | 5.2 | No |
| 3775 | 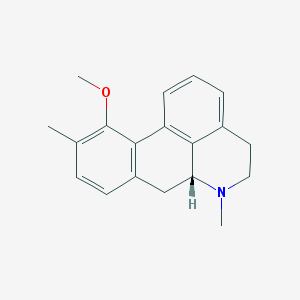 CHEMBL332178 CHEMBL332178 | C19H21NO | 279.383 | 2 / 0 | 3.3 | Yes |
| 4510 |  CHEMBL497604 CHEMBL497604 | C24H26N4O2 | 402.498 | 5 / 1 | 4.0 | Yes |
| 4619 |  CHEMBL95112 CHEMBL95112 | C23H27FN2O2S | 414.539 | 6 / 0 | 3.4 | Yes |
| 4978 |  CHEMBL190525 CHEMBL190525 | C26H29N5O3 | 459.55 | 5 / 2 | 3.4 | Yes |
| 5454 |  CHEMBL175226 CHEMBL175226 | C19H24ClN3OS | 377.931 | 4 / 0 | 2.8 | Yes |
| 5550 | 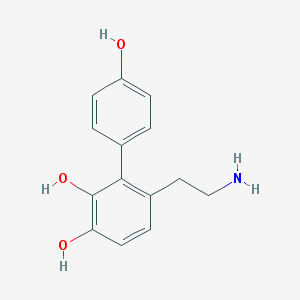 CHEMBL57470 CHEMBL57470 | C14H15NO3 | 245.278 | 4 / 4 | 1.8 | Yes |
| 5713 | 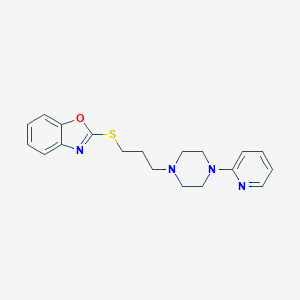 CHEMBL517962 CHEMBL517962 | C19H22N4OS | 354.472 | 6 / 0 | 3.9 | Yes |
| 5758 | 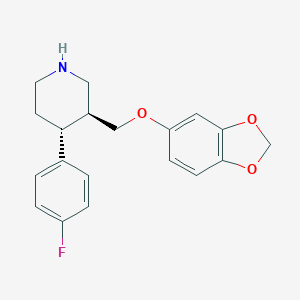 paroxetine paroxetine | C19H20FNO3 | 329.371 | 5 / 1 | 3.5 | Yes |
| 6871 | 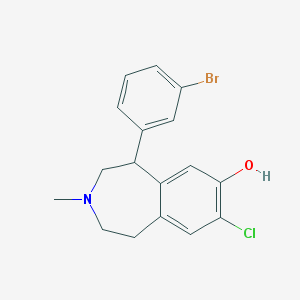 CHEMBL13632 CHEMBL13632 | C17H17BrClNO | 366.683 | 2 / 1 | 4.7 | Yes |
| 6880 |  CHEMBL171519 CHEMBL171519 | C14H15N3 | 225.295 | 2 / 2 | 1.5 | Yes |
| 7244 |  CHEMBL426813 CHEMBL426813 | C21H24ClN3O3 | 401.891 | 5 / 0 | 2.6 | Yes |
| 7312 |  CHEMBL289557 CHEMBL289557 | C16H24ClNO | 281.824 | 2 / 1 | 4.7 | Yes |
| 7314 |  CHEMBL543613 CHEMBL543613 | C22H24ClFN2O2S | 434.954 | 6 / 1 | N/A | N/A |
| 7692 | 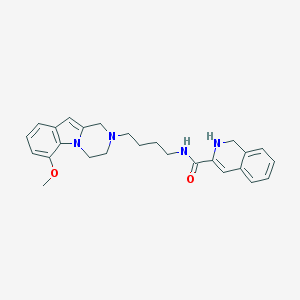 CID 44581146 CID 44581146 | C26H30N4O2 | 430.552 | 4 / 2 | 3.5 | Yes |
| 7870 | 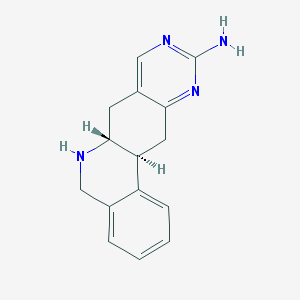 CHEMBL173658 CHEMBL173658 | C15H16N4 | 252.321 | 4 / 2 | 1.1 | Yes |
| 7964 | 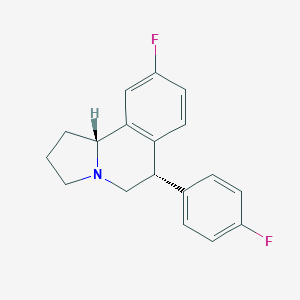 CHEMBL1743798 CHEMBL1743798 | C18H17F2N | 285.338 | 3 / 0 | 4.0 | Yes |
| 8119 | 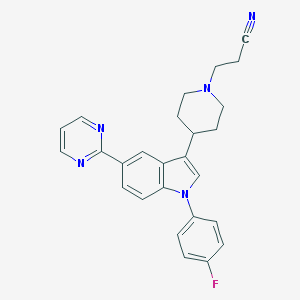 CHEMBL317333 CHEMBL317333 | C26H24FN5 | 425.511 | 5 / 0 | 4.0 | Yes |
| 8193 |  CHEMBL1743781 CHEMBL1743781 | C18H20BrNO | 346.268 | 2 / 2 | N/A | N/A |
| 8205 |  BW-723C86 BW-723C86 | C16H18N2OS | 286.393 | 3 / 2 | 3.0 | Yes |
| 9134 |  CHEMBL542167 CHEMBL542167 | C19H24ClN | 301.858 | 1 / 1 | N/A | N/A |
| 11179 |  ISOCLOZAPINE ISOCLOZAPINE | C18H19ClN4 | 326.828 | 3 / 1 | 3.1 | Yes |
| 11398 | 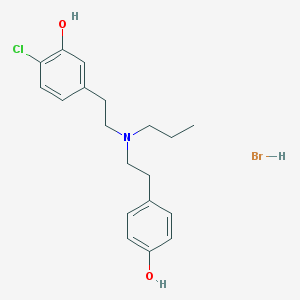 CHEMBL555427 CHEMBL555427 | C19H25BrClNO2 | 414.768 | 3 / 3 | N/A | N/A |
| 11865 | 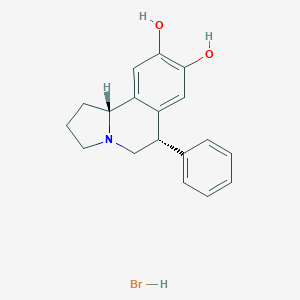 CHEMBL1743799 CHEMBL1743799 | C18H20BrNO2 | 362.267 | 3 / 3 | N/A | N/A |
| 11867 | 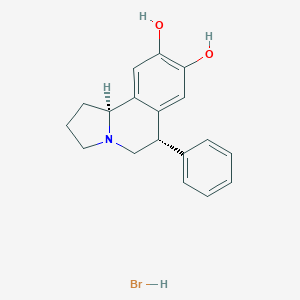 CHEMBL1743774 CHEMBL1743774 | C18H20BrNO2 | 362.267 | 3 / 3 | N/A | N/A |
| 12431 | 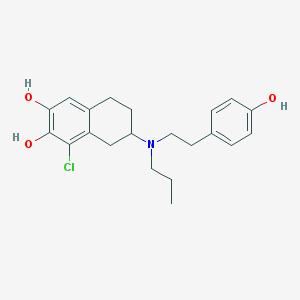 CHEMBL40949 CHEMBL40949 | C21H26ClNO3 | 375.893 | 4 / 3 | 5.1 | No |
| 12478 | 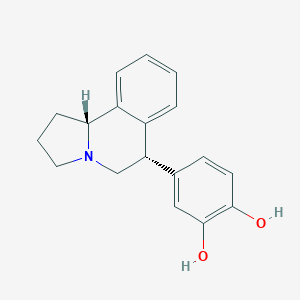 CHEMBL1743819 CHEMBL1743819 | C18H19NO2 | 281.355 | 3 / 2 | 3.1 | Yes |
| 13195 | 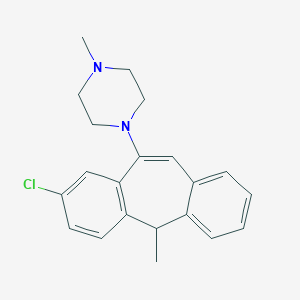 CHEMBL72220 CHEMBL72220 | C21H23ClN2 | 338.879 | 2 / 0 | 4.9 | Yes |
| 13622 | 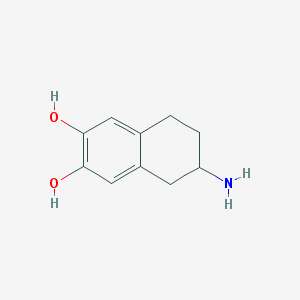 ADTN ADTN | C10H13NO2 | 179.219 | 3 / 3 | 1.4 | Yes |
| 13687 | 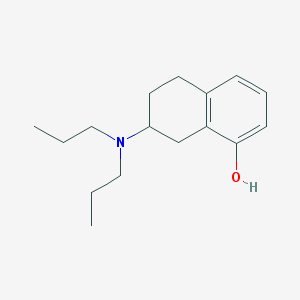 8-OH-Dpat 8-OH-Dpat | C16H25NO | 247.382 | 2 / 1 | 4.1 | Yes |
| 13697 | 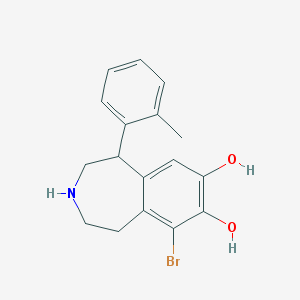 BDBM86280 BDBM86280 | C17H18BrNO2 | 348.24 | 3 / 3 | 3.2 | Yes |
| 13745 | 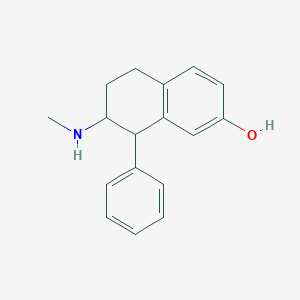 CHEMBL111083 CHEMBL111083 | C17H19NO | 253.345 | 2 / 2 | 3.3 | Yes |
| 14412 | 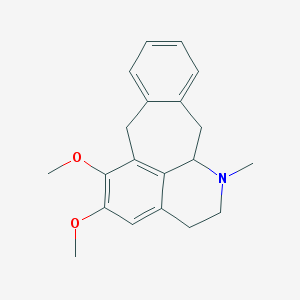 CHEMBL404240 CHEMBL404240 | C20H23NO2 | 309.409 | 3 / 0 | 3.7 | Yes |
| 15016 | 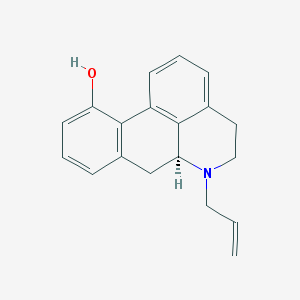 CHEMBL1744043 CHEMBL1744043 | C19H19NO | 277.367 | 2 / 1 | 3.3 | Yes |
| 15018 | 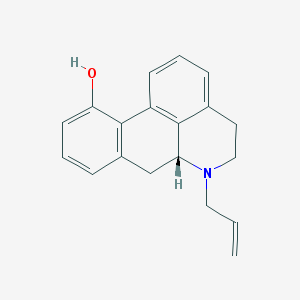 CHEMBL1788212 CHEMBL1788212 | C19H19NO | 277.367 | 2 / 1 | 3.3 | Yes |
| 15453 | 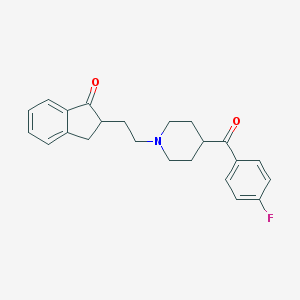 CHEMBL87717 CHEMBL87717 | C23H24FNO2 | 365.448 | 4 / 0 | 4.0 | Yes |
| 15729 | 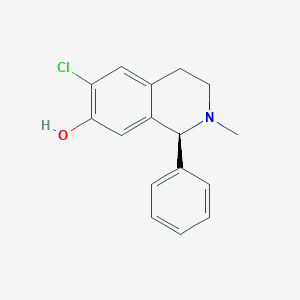 CHEMBL303993 CHEMBL303993 | C16H16ClNO | 273.76 | 2 / 1 | 3.8 | Yes |
| 15731 | 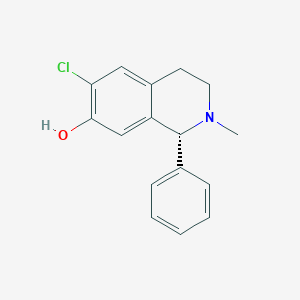 CHEMBL293158 CHEMBL293158 | C16H16ClNO | 273.76 | 2 / 1 | 3.8 | Yes |
| 15736 | 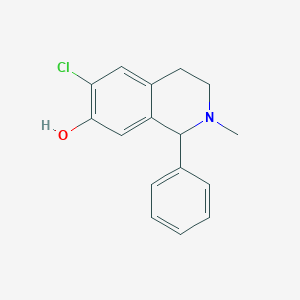 CHEMBL57579 CHEMBL57579 | C16H16ClNO | 273.76 | 2 / 1 | 3.8 | Yes |
| 15893 | 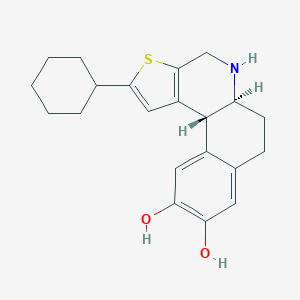 CHEMBL290149 CHEMBL290149 | C21H25NO2S | 355.496 | 4 / 3 | 4.4 | Yes |
| 16082 | 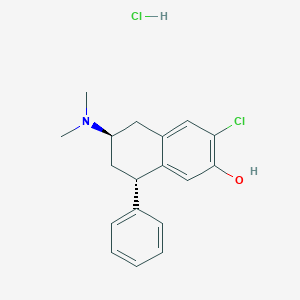 CHEMBL545477 CHEMBL545477 | C18H21Cl2NO | 338.272 | 2 / 2 | N/A | N/A |
| 16241 | 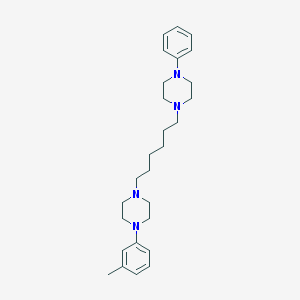 CHEMBL1172734 CHEMBL1172734 | C27H40N4 | 420.645 | 4 / 0 | 5.3 | No |
| 16406 | 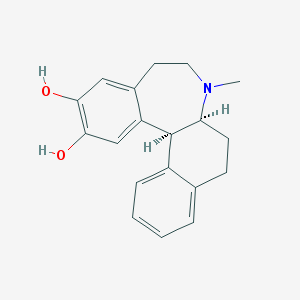 CHEMBL301003 CHEMBL301003 | C19H21NO2 | 295.382 | 3 / 2 | 3.5 | Yes |
| 16408 | 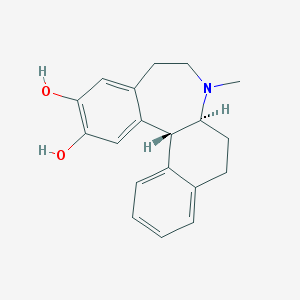 CHEMBL56100 CHEMBL56100 | C19H21NO2 | 295.382 | 3 / 2 | 3.5 | Yes |
| 16702 | 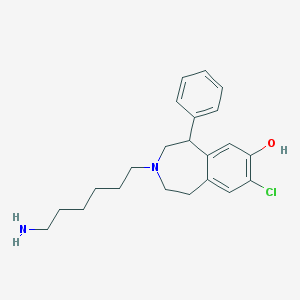 CHEMBL137637 CHEMBL137637 | C22H29ClN2O | 372.937 | 3 / 2 | 4.5 | Yes |
| 16799 | 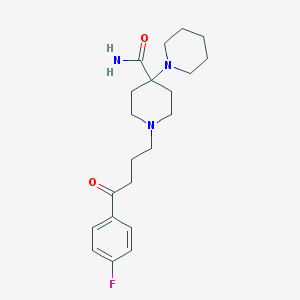 Pipamperone Pipamperone | C21H30FN3O2 | 375.488 | 5 / 1 | 2.0 | Yes |
| 18089 | 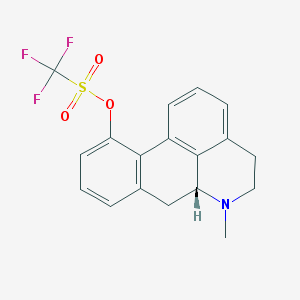 CHEMBL333836 CHEMBL333836 | C18H16F3NO3S | 383.385 | 7 / 0 | 3.8 | Yes |
| 18220 | 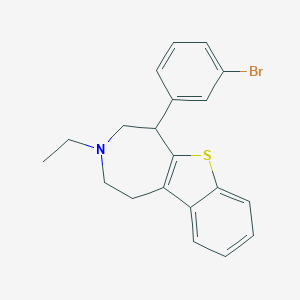 CHEMBL139081 CHEMBL139081 | C20H20BrNS | 386.351 | 2 / 0 | 5.8 | No |
| 19397 | 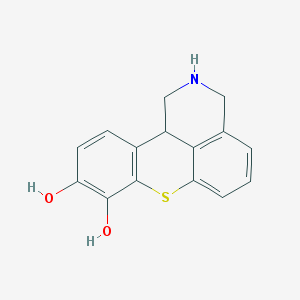 BDBM86461 BDBM86461 | C15H13NO2S | 271.334 | 4 / 3 | 2.2 | Yes |
| 19405 | 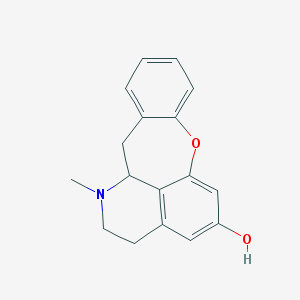 CHEMBL155208 CHEMBL155208 | C17H17NO2 | 267.328 | 3 / 1 | 3.0 | Yes |
| 20173 |  imipramine imipramine | C19H24N2 | 280.415 | 2 / 0 | 4.8 | Yes |
| 522065 |  CHEMBL3763812 CHEMBL3763812 | C18H21FN4O | 328.391 | 5 / 1 | 1.9 | Yes |
| 20616 |  CHEMBL192037 CHEMBL192037 | C14H24N2 | 220.36 | 1 / 0 | 2.9 | Yes |
| 20761 |  CHEMBL67355 CHEMBL67355 | C17H19N5 | 293.374 | 4 / 1 | 1.7 | Yes |
| 522072 | 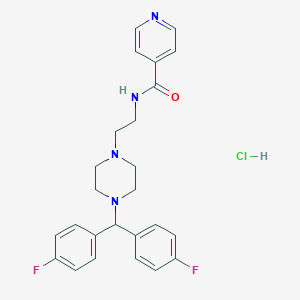 CHEMBL3765471 CHEMBL3765471 | C25H27ClF2N4O | 472.965 | 6 / 2 | N/A | N/A |
| 21233 | 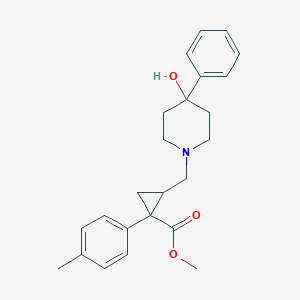 (RS)-PPCC (RS)-PPCC | C24H29NO3 | 379.5 | 4 / 1 | 3.5 | Yes |
| 21416 | 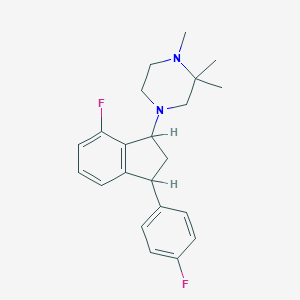 CHEMBL137536 CHEMBL137536 | C22H26F2N2 | 356.461 | 4 / 0 | 4.3 | Yes |
| 21595 | 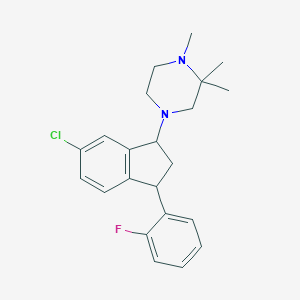 CHEMBL136468 CHEMBL136468 | C22H26ClFN2 | 372.912 | 3 / 0 | 4.8 | Yes |
| 21608 |  CHEMBL543964 CHEMBL543964 | C16H16ClN3O | 301.774 | 1 / 3 | N/A | N/A |
| 22321 |  CHEMBL363466 CHEMBL363466 | C20H25NO2 | 311.425 | 3 / 0 | 4.2 | Yes |
| 22542 |  CHEMBL283036 CHEMBL283036 | C19H19F3N4 | 360.384 | 6 / 1 | 3.6 | Yes |
| 22894 |  CHEMBL85879 CHEMBL85879 | C20H26N2 | 294.442 | 2 / 0 | 4.5 | Yes |
| 23227 | 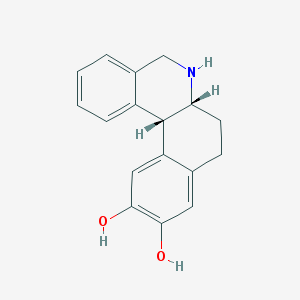 CHEMBL43345 CHEMBL43345 | C17H17NO2 | 267.328 | 3 / 3 | 2.5 | Yes |
| 23237 | 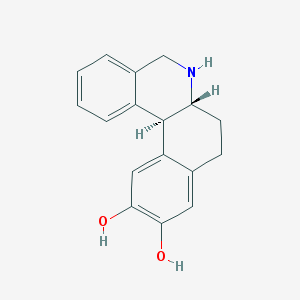 DIHYDREXIDINE DIHYDREXIDINE | C17H17NO2 | 267.328 | 3 / 3 | 2.5 | Yes |
| 23250 | 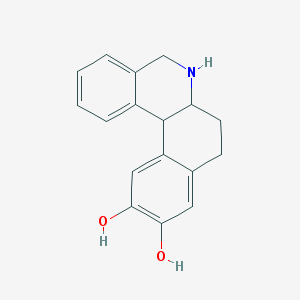 DIHYDREXIDINE DIHYDREXIDINE | C17H17NO2 | 267.328 | 3 / 3 | 2.5 | Yes |
| 553360 | 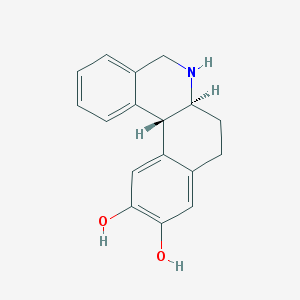 DIHYDREXIDINE DIHYDREXIDINE | C17H17NO2 | 267.328 | 3 / 3 | 2.5 | Yes |
| 23330 | 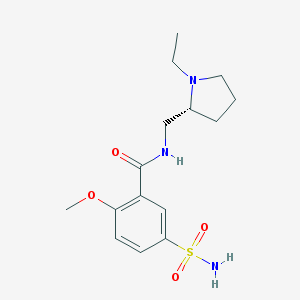 (+)-sulpiride (+)-sulpiride | C15H23N3O4S | 341.426 | 6 / 2 | 0.6 | Yes |
| 23346 | 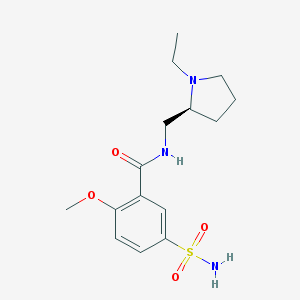 Levosulpiride Levosulpiride | C15H23N3O4S | 341.426 | 6 / 2 | 0.6 | Yes |
| 23348 | 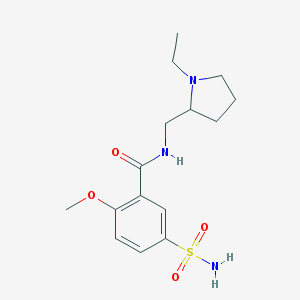 sulpiride sulpiride | C15H23N3O4S | 341.426 | 6 / 2 | 0.6 | Yes |
| 24477 | 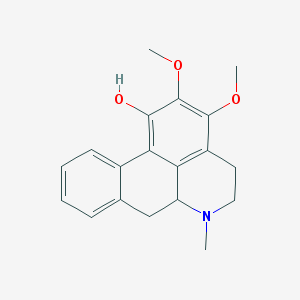 CHEMBL257747 CHEMBL257747 | C19H21NO3 | 311.381 | 4 / 1 | 3.1 | Yes |
| 24768 | 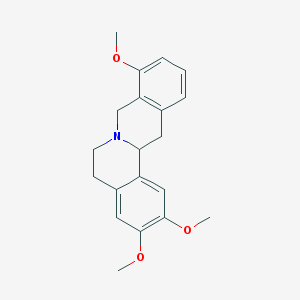 SCHEMBL910344 SCHEMBL910344 | C20H23NO3 | 325.408 | 4 / 0 | 3.3 | Yes |
| 25012 | 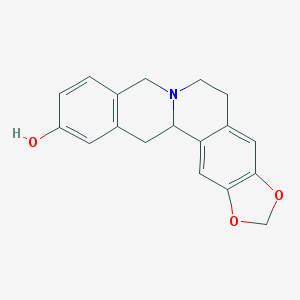 CHEMBL2425377 CHEMBL2425377 | C18H17NO3 | 295.338 | 4 / 1 | 2.8 | Yes |
| 25146 | 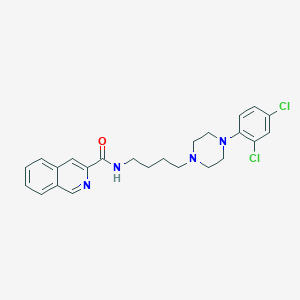 CHEMBL340641 CHEMBL340641 | C24H26Cl2N4O | 457.399 | 4 / 1 | 5.3 | No |
| 25499 | 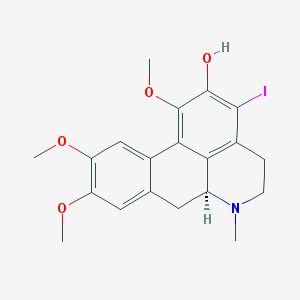 CHEMBL401690 CHEMBL401690 | C20H22INO4 | 467.303 | 5 / 1 | 3.7 | Yes |
| 25790 | 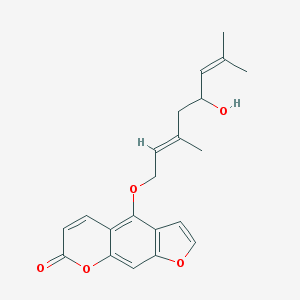 Notopterol Notopterol | C21H22O5 | 354.402 | 5 / 1 | 4.5 | Yes |
| 28265 | 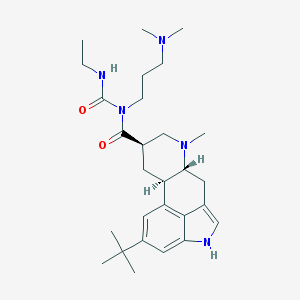 CHEMBL276013 CHEMBL276013 | C28H43N5O2 | 481.685 | 4 / 2 | 4.4 | Yes |
| 28454 | 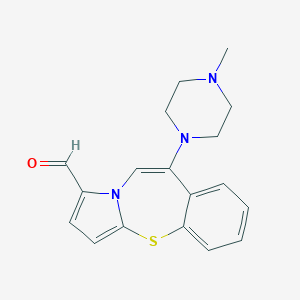 CHEMBL362060 CHEMBL362060 | C18H19N3OS | 325.43 | 4 / 0 | 2.7 | Yes |
| 28686 | 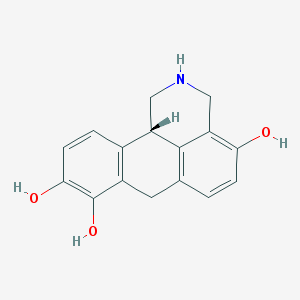 4-hydroxydinapsoline hydrobromide 4-hydroxydinapsoline hydrobromide | C16H15NO3 | 269.3 | 4 / 4 | 1.7 | Yes |
| 28759 | 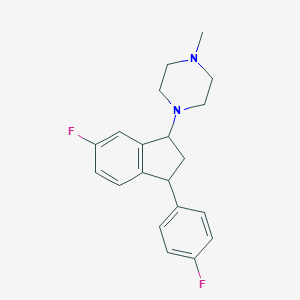 CHEMBL307717 CHEMBL307717 | C20H22F2N2 | 328.407 | 4 / 0 | 3.6 | Yes |
| 28858 | 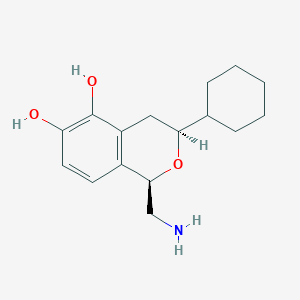 CHEMBL100572 CHEMBL100572 | C16H23NO3 | 277.364 | 4 / 3 | 2.4 | Yes |
| 28860 | 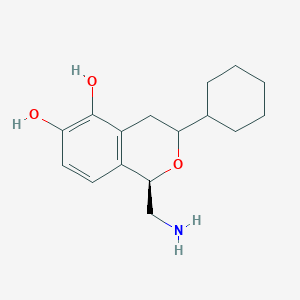 CHEMBL314459 CHEMBL314459 | C16H23NO3 | 277.364 | 4 / 3 | 2.4 | Yes |
| 30167 | 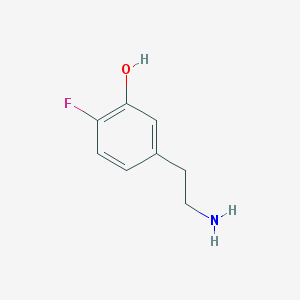 CHEMBL545240 CHEMBL545240 | C8H10FNO | 155.172 | 3 / 2 | -0.5 | Yes |
| 31144 | 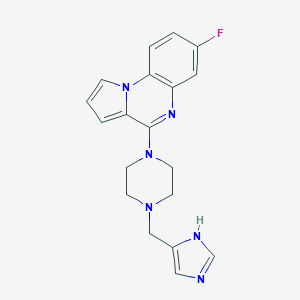 CHEMBL566522 CHEMBL566522 | C19H19FN6 | 350.401 | 5 / 1 | 2.9 | Yes |
| 31717 | 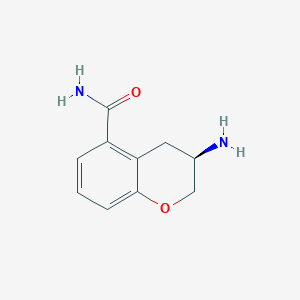 CHEMBL2112388 CHEMBL2112388 | C10H12N2O2 | 192.218 | 3 / 2 | -0.1 | Yes |
| 32059 | 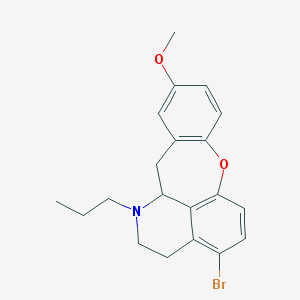 CHEMBL349311 CHEMBL349311 | C20H22BrNO2 | 388.305 | 3 / 0 | 4.9 | Yes |
| 32228 | 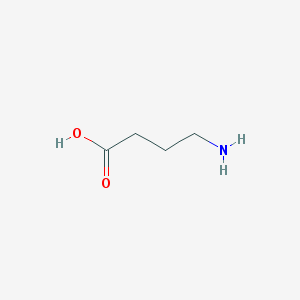 4-aminobutyric acid 4-aminobutyric acid | C4H9NO2 | 103.121 | 3 / 2 | -3.2 | Yes |
| 32308 |  CHEMBL330274 CHEMBL330274 | C19H21NO2 | 295.382 | 3 / 2 | 3.2 | Yes |
| 32311 |  n-Propylapomorphine n-Propylapomorphine | C19H21NO2 | 295.382 | 3 / 2 | 3.2 | Yes |
| 32428 |  CHEMBL41809 CHEMBL41809 | C20H25NO2S | 343.485 | 4 / 3 | 4.2 | Yes |
| 33304 |  BDBM50265778 BDBM50265778 | C26H30N4O2 | 430.552 | 4 / 2 | 3.1 | Yes |
zhanglab![]() zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218