

You can:
| Name | P2Y purinoceptor 6 |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | P2RY6 |
| Synonym | pyrimidinergic receptor P2Y P2Y6 receptor P2Y6 P2Y purinoceptor 6 P2Y ATP receptor 6 [ Show all ] |
| Disease | N/A |
| Length | 328 |
| Amino acid sequence | MEWDNGTGQALGLPPTTCVYRENFKQLLLPPVYSAVLAAGLPLNICVITQICTSRRALTRTAVYTLNLALADLLYACSLPLLIYNYAQGDHWPFGDFACRLVRFLFYANLHGSILFLTCISFQRYLGICHPLAPWHKRGGRRAAWLVCVAVWLAVTTQCLPTAIFAATGIQRNRTVCYDLSPPALATHYMPYGMALTVIGFLLPFAALLACYCLLACRLCRQDGPAEPVAQERRGKAARMAVVVAAAFAISFLPFHITKTAYLAVRSTPGVPCTVLEAFAAAYKGTRPFASANSVLDPILFYFTQKKFRRRPHELLQKLTAKWQRQGR |
| UniProt | Q15077 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | Q15077 |
| 3D structure model | This predicted structure model is from GPCR-EXP Q15077. |
| BioLiP | N/A |
| Therapeutic Target Database | N/A |
| ChEMBL | CHEMBL4714 |
| IUPHAR | 326 |
| DrugBank | N/A |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 3491 | 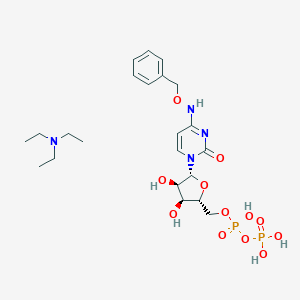 CHEMBL1084612 CHEMBL1084612 | C22H36N4O12P2 | 610.494 | 13 / 6 | N/A | No |
| 4531 | 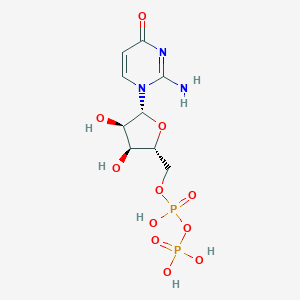 CHEMBL1767415 CHEMBL1767415 | C9H15N3O11P2 | 403.177 | 11 / 6 | -4.5 | No |
| 4855 | 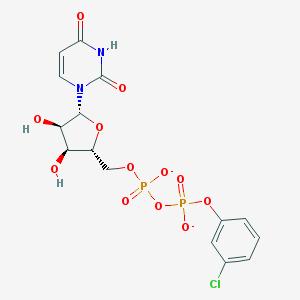 BDBM50303350 BDBM50303350 | C15H15ClN2O12P2-2 | 512.685 | 12 / 3 | -1.9 | No |
| 5498 | 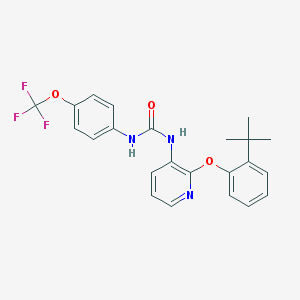 BPTU BPTU | C23H22F3N3O3 | 445.442 | 7 / 2 | 6.1 | No |
| 6218 | 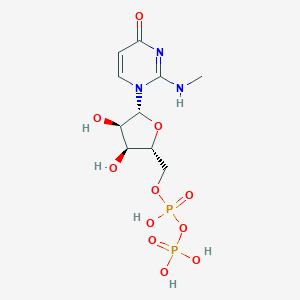 CHEMBL1767416 CHEMBL1767416 | C10H17N3O11P2 | 417.204 | 11 / 6 | -4.1 | No |
| 7016 | 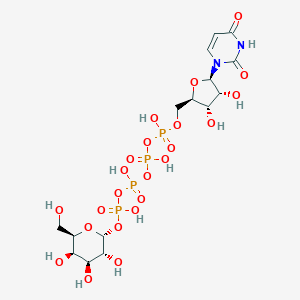 CHEMBL444212 CHEMBL444212 | C15H26N2O23P4 | 726.259 | 23 / 11 | -8.5 | No |
| 7018 | 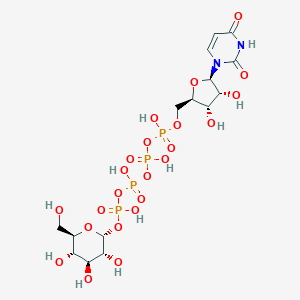 CHEMBL499138 CHEMBL499138 | C15H26N2O23P4 | 726.259 | 23 / 11 | -8.5 | No |
| 7022 | 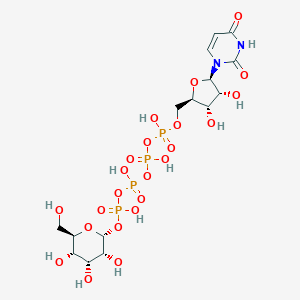 CHEMBL1784885 CHEMBL1784885 | C15H26N2O23P4 | 726.259 | 23 / 11 | -8.5 | No |
| 7023 | 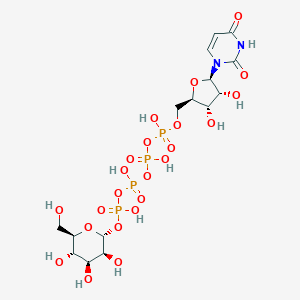 CHEMBL1784886 CHEMBL1784886 | C15H26N2O23P4 | 726.259 | 23 / 11 | -8.5 | No |
| 10834 | 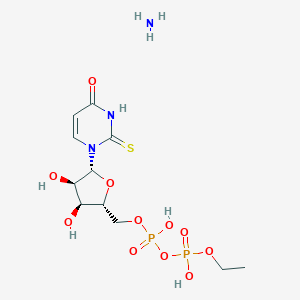 CHEMBL1162563 CHEMBL1162563 | C11H21N3O11P2S | 465.307 | 13 / 6 | N/A | No |
| 12535 | 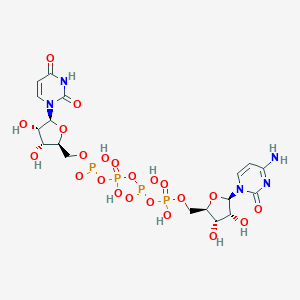 CHEMBL1162198 CHEMBL1162198 | C18H27N5O22P4 | 789.322 | 22 / 10 | -9.4 | No |
| 12611 | 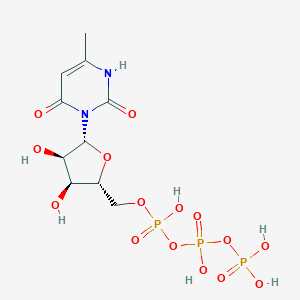 CHEMBL413841 CHEMBL413841 | C10H17N2O15P3 | 498.166 | 15 / 7 | -5.8 | No |
| 15818 | 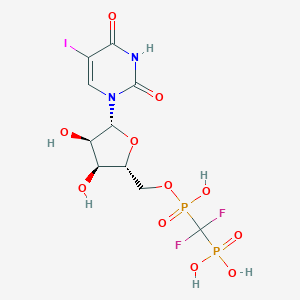 CHEMBL566925 CHEMBL566925 | C10H13F2IN2O11P2 | 564.066 | 13 / 6 | -3.8 | No |
| 16609 | 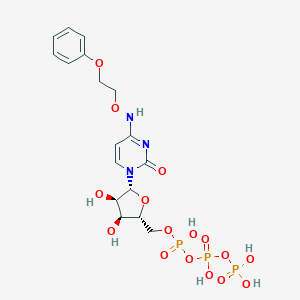 CHEMBL3261361 CHEMBL3261361 | C17H24N3O16P3 | 619.305 | 16 / 7 | -3.3 | No |
| 17594 | 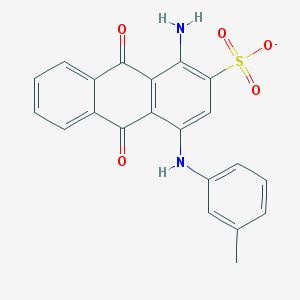 CHEMBL445413 CHEMBL445413 | C21H15N2O5S- | 407.42 | 7 / 2 | 3.9 | Yes |
| 18302 | 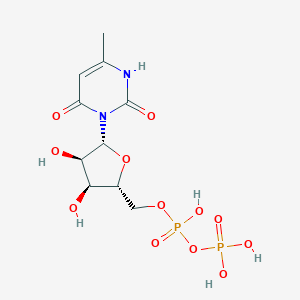 CHEMBL386096 CHEMBL386096 | C10H16N2O12P2 | 418.188 | 12 / 6 | -4.7 | No |
| 18897 | 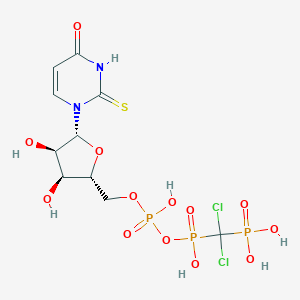 CHEMBL519835 CHEMBL519835 | C10H15Cl2N2O13P3S | 567.112 | 14 / 7 | -4.4 | No |
| 19506 | 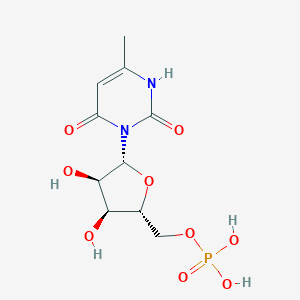 CHEMBL215263 CHEMBL215263 | C10H15N2O9P | 338.209 | 9 / 5 | -3.6 | Yes |
| 21438 | 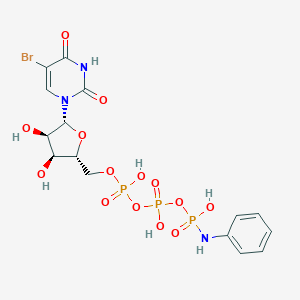 CHEMBL1765120 CHEMBL1765120 | C15H19BrN3O14P3 | 638.149 | 15 / 7 | -2.3 | No |
| 22714 | 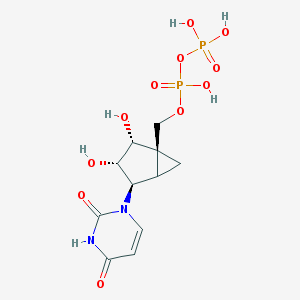 CHEMBL215811 CHEMBL215811 | C11H16N2O11P2 | 414.2 | 11 / 6 | -4.6 | No |
| 24068 | 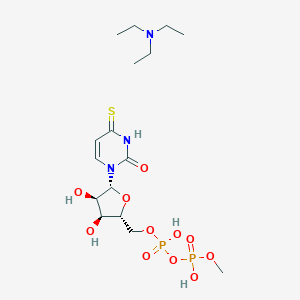 CHEMBL608851 CHEMBL608851 | C16H31N3O11P2S | 535.442 | 13 / 5 | N/A | No |
| 26411 | 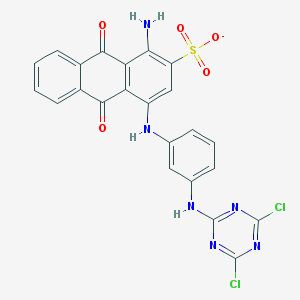 CHEMBL1672105 CHEMBL1672105 | C23H13Cl2N6O5S- | 556.354 | 11 / 3 | 5.3 | No |
| 27555 | 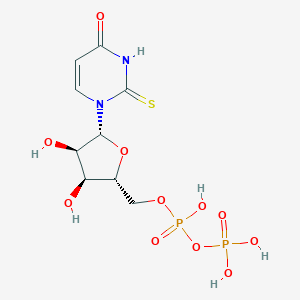 2-thio-UDP 2-thio-UDP | C9H14N2O11P2S | 420.222 | 12 / 6 | -4.1 | No |
| 30314 | 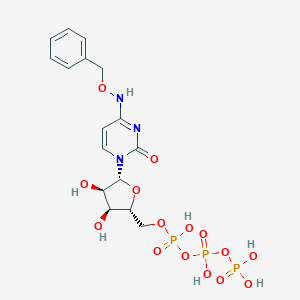 CHEMBL1784892 CHEMBL1784892 | C16H22N3O15P3 | 589.279 | 15 / 7 | -3.7 | No |
| 31387 | 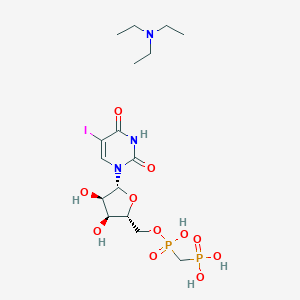 CHEMBL1085561 CHEMBL1085561 | C16H30IN3O11P2 | 629.278 | 12 / 6 | N/A | No |
| 32088 | 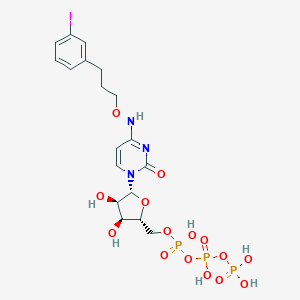 CHEMBL3261365 CHEMBL3261365 | C18H25IN3O15P3 | 743.23 | 15 / 7 | -3.1 | No |
| 32486 | 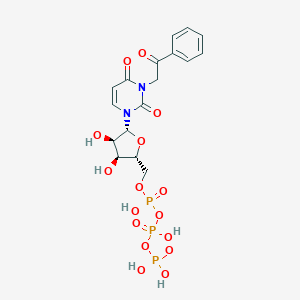 CHEMBL385488 CHEMBL385488 | C17H21N2O16P3 | 602.274 | 16 / 6 | -4.5 | No |
| 33910 | 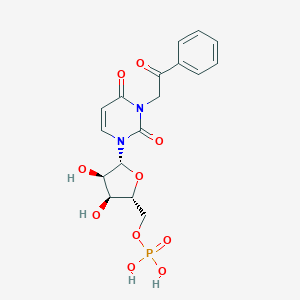 CHEMBL215381 CHEMBL215381 | C17H19N2O10P | 442.317 | 10 / 4 | -2.4 | Yes |
| 34045 | 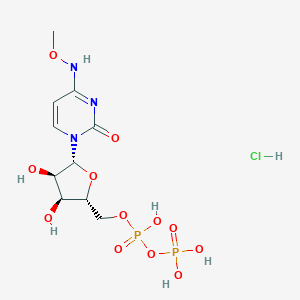 CHEMBL1204010 CHEMBL1204010 | C10H18ClN3O12P2 | 469.661 | 12 / 7 | N/A | No |
| 37982 | 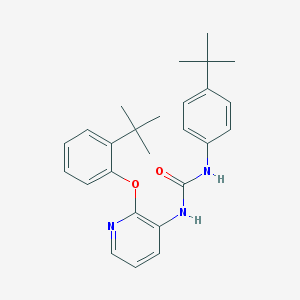 CHEMBL2333771 CHEMBL2333771 | C26H31N3O2 | 417.553 | 3 / 2 | 6.6 | No |
| 38297 | 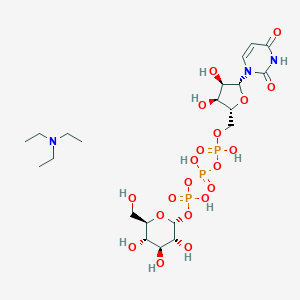 CHEMBL1083260 CHEMBL1083260 | C21H40N3O20P3 | 747.473 | 21 / 10 | N/A | No |
| 38816 | 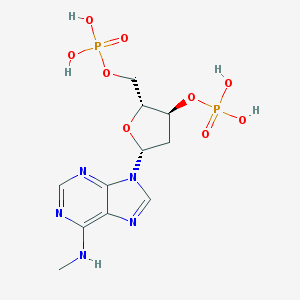 CHEMBL129841 CHEMBL129841 | C11H17N5O9P2 | 425.231 | 13 / 5 | -2.9 | No |
| 42143 | 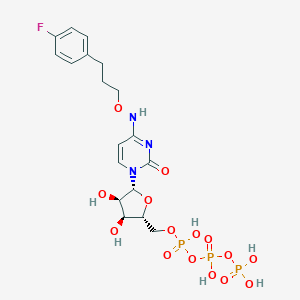 CHEMBL3261363 CHEMBL3261363 | C18H25FN3O15P3 | 635.324 | 16 / 7 | -3.6 | No |
| 42246 | 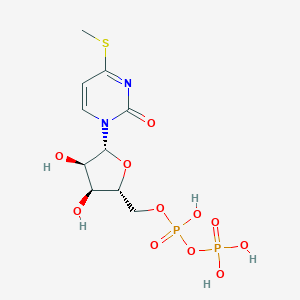 CHEMBL1199698 CHEMBL1199698 | C10H16N2O11P2S | 434.249 | 12 / 5 | -3.0 | No |
| 42289 | 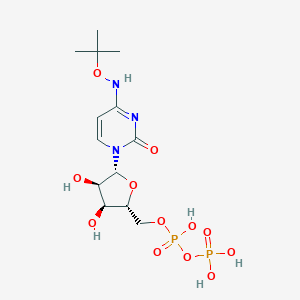 CHEMBL1084296 CHEMBL1084296 | C13H23N3O12P2 | 475.284 | 12 / 6 | -3.1 | No |
| 44382 | 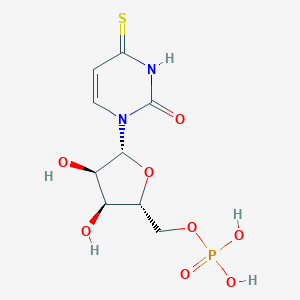 CHEMBL1230331 CHEMBL1230331 | C9H13N2O8PS | 340.243 | 9 / 5 | -3.0 | Yes |
| 47051 | 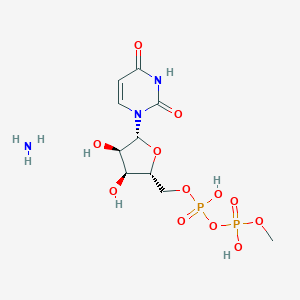 CHEMBL1162554 CHEMBL1162554 | C10H19N3O12P2 | 435.219 | 13 / 6 | N/A | No |
| 47983 | 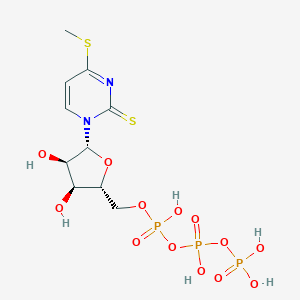 CHEMBL482472 CHEMBL482472 | C10H17N2O13P3S2 | 530.288 | 15 / 6 | -4.3 | No |
| 49428 | 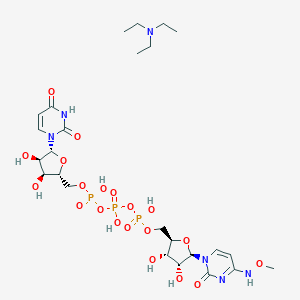 CHEMBL1083256 CHEMBL1083256 | C25H43N6O20P3 | 840.562 | 21 / 9 | N/A | No |
| 49771 | 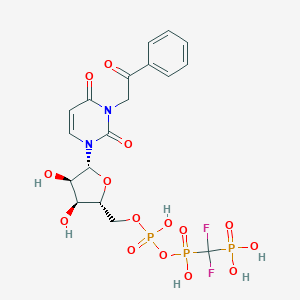 CHEMBL1765118 CHEMBL1765118 | C18H21F2N2O15P3 | 636.283 | 17 / 6 | -3.9 | No |
| 53514 | 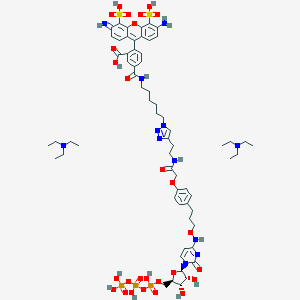 CHEMBL3261377 CHEMBL3261377 | C63H89N12O27P3S2 | 1603.5 | 33 / 14 | N/A | No |
| 53630 | 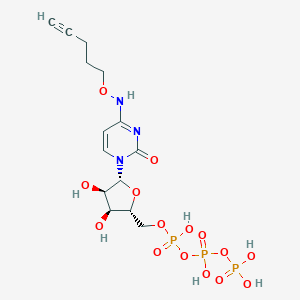 CHEMBL3261373 CHEMBL3261373 | C14H22N3O15P3 | 565.257 | 15 / 7 | -4.2 | No |
| 58256 | 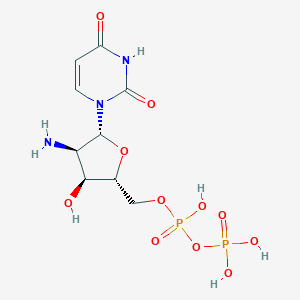 CHEMBL215911 CHEMBL215911 | C9H15N3O11P2 | 403.177 | 12 / 6 | -7.3 | No |
| 58528 | 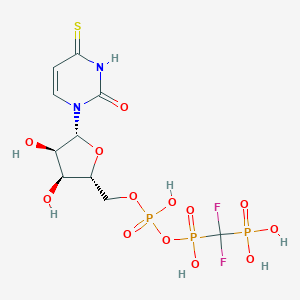 CHEMBL1765119 CHEMBL1765119 | C10H15F2N2O13P3S | 534.209 | 16 / 7 | -4.6 | No |
| 60004 | 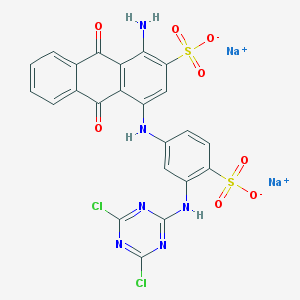 Azure KX Azure KX | C23H12Cl2N6Na2O8S2 | 681.383 | 14 / 3 | N/A | No |
| 62393 | 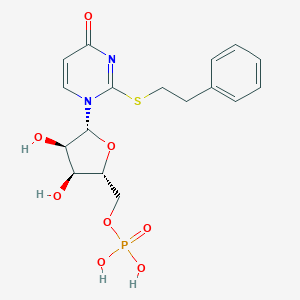 CHEMBL1767421 CHEMBL1767421 | C17H21N2O8PS | 444.395 | 9 / 4 | 0.1 | Yes |
| 66142 | 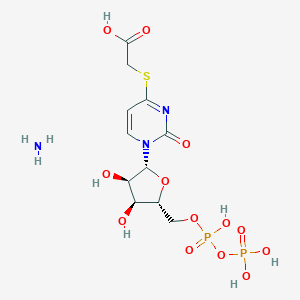 CHEMBL1161885 CHEMBL1161885 | C11H19N3O13P2S | 495.289 | 15 / 7 | N/A | No |
| 68553 | 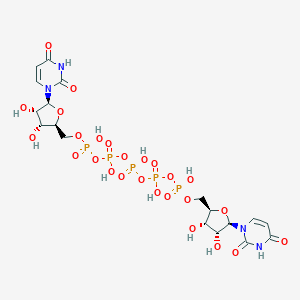 CHEMBL2113401 CHEMBL2113401 | C18H27N4O26P5 | 870.285 | 26 / 11 | -10.2 | No |
| 68640 |  CHEMBL1083263 CHEMBL1083263 | C22H42N3O19P3 | 745.501 | 20 / 10 | N/A | No |
| 69885 |  CHEMBL1083258 CHEMBL1083258 | C15H25N2O15P3 | 566.285 | 15 / 6 | -3.5 | No |
| 72313 |  CHEMBL219779 CHEMBL219779 | C9H14N5O15P3 | 525.152 | 17 / 7 | -5.0 | No |
| 72459 |  CHEMBL1083264 CHEMBL1083264 | C22H43N4O20P3 | 776.515 | 21 / 10 | N/A | No |
| 73199 | 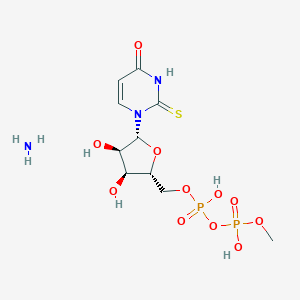 CHEMBL1162557 CHEMBL1162557 | C10H19N3O11P2S | 451.28 | 13 / 6 | N/A | No |
| 73966 | 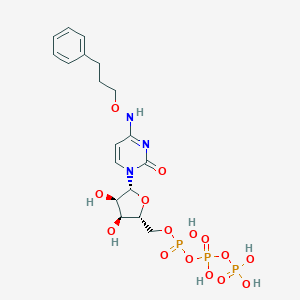 MRS-4062 MRS-4062 | C18H26N3O15P3 | 617.333 | 15 / 7 | -3.7 | No |
| 74632 | 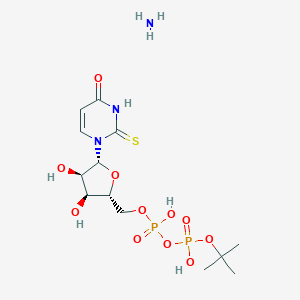 CHEMBL1162556 CHEMBL1162556 | C13H25N3O11P2S | 493.361 | 13 / 6 | N/A | No |
| 75092 | 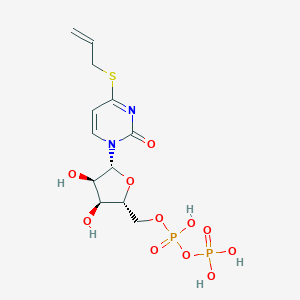 CHEMBL1199952 CHEMBL1199952 | C12H18N2O11P2S | 460.287 | 12 / 5 | -2.4 | No |
| 75412 | 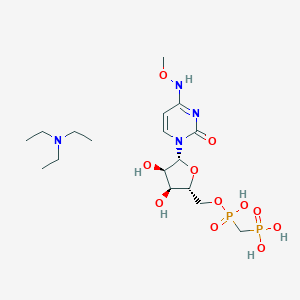 CHEMBL1084613 CHEMBL1084613 | C17H34N4O11P2 | 532.424 | 12 / 6 | N/A | No |
| 75982 | 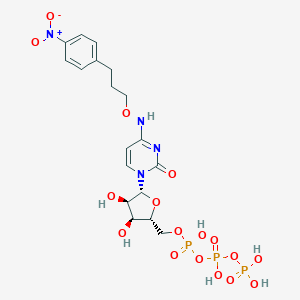 CHEMBL3261368 CHEMBL3261368 | C18H25N4O17P3 | 662.33 | 17 / 7 | -3.9 | No |
| 76101 | 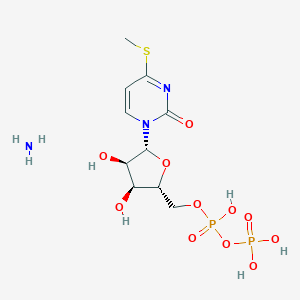 CHEMBL1161878 CHEMBL1161878 | C10H19N3O11P2S | 451.28 | 13 / 6 | N/A | No |
| 76439 | 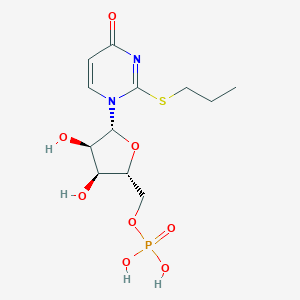 CHEMBL1767419 CHEMBL1767419 | C12H19N2O8PS | 382.324 | 9 / 4 | -1.0 | Yes |
| 559691 | 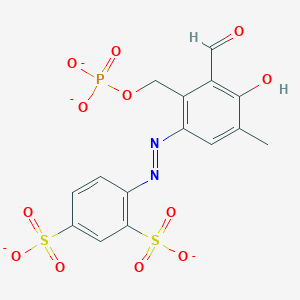 CHEMBL477339 CHEMBL477339 | C15H11N2O12PS2-4 | 506.349 | 14 / 1 | -1.1 | No |
| 77911 | 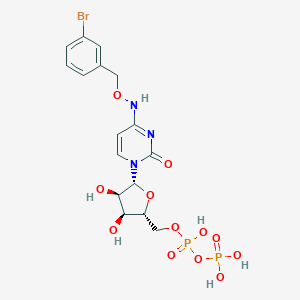 CHEMBL3220046 CHEMBL3220046 | C16H20BrN3O12P2 | 588.197 | 12 / 6 | -2.7 | No |
| 78870 | 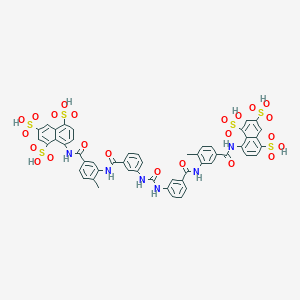 suramin suramin | C51H40N6O23S6 | 1297.26 | 23 / 12 | 1.5 | No |
| 79522 | 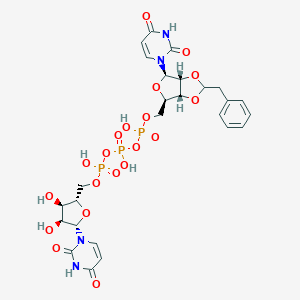 CHEMBL1162174 CHEMBL1162174 | C26H31N4O20P3 | 812.463 | 20 / 7 | -5.0 | No |
| 79697 | 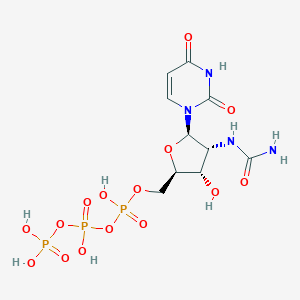 CHEMBL484295 CHEMBL484295 | C10H17N4O15P3 | 526.18 | 15 / 8 | -6.4 | No |
| 81348 | 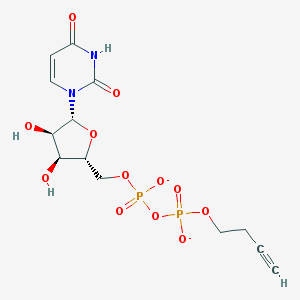 BDBM50303345 BDBM50303345 | C13H16N2O12P2-2 | 454.221 | 12 / 3 | -3.8 | No |
| 81559 | 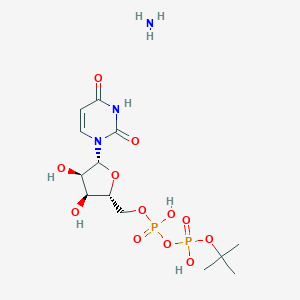 CHEMBL1162552 CHEMBL1162552 | C13H25N3O12P2 | 477.3 | 13 / 6 | N/A | No |
| 82498 | 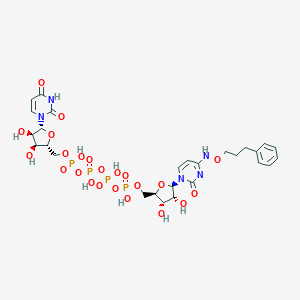 CHEMBL3261379 CHEMBL3261379 | C27H37N5O23P4 | 923.5 | 23 / 10 | -6.7 | No |
| 83660 | 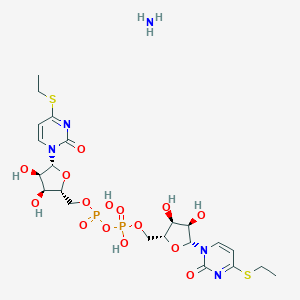 CHEMBL1161872 CHEMBL1161872 | C22H35N5O15P2S2 | 735.61 | 18 / 7 | N/A | No |
| 83772 | 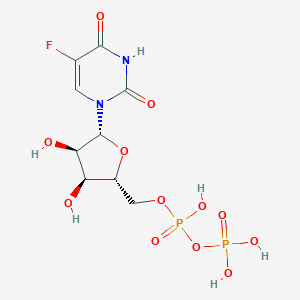 5-fluorouridine diphosphate 5-fluorouridine diphosphate | C9H13FN2O12P2 | 422.151 | 13 / 6 | -4.7 | No |
| 84172 | 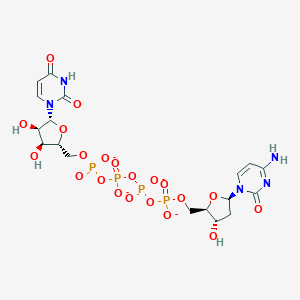 CHEMBL1767407 CHEMBL1767407 | C18H23N5O21P4-4 | 769.291 | 21 / 5 | -8.8 | No |
| 84173 | 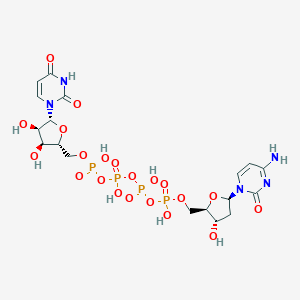 denufosol denufosol | C18H27N5O21P4 | 773.323 | 21 / 9 | -8.4 | No |
| 85064 | 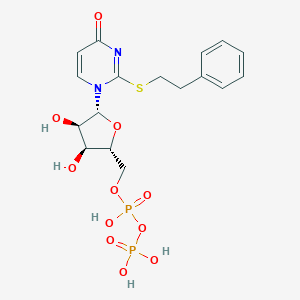 CHEMBL1767413 CHEMBL1767413 | C17H22N2O11P2S | 524.374 | 12 / 5 | -1.9 | No |
| 87914 | 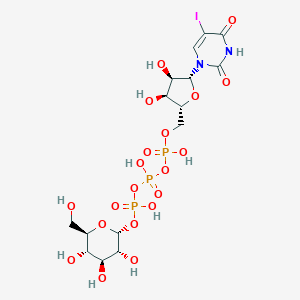 CHEMBL1083261 CHEMBL1083261 | C15H24IN2O20P3 | 772.177 | 20 / 10 | -7.1 | No |
| 88249 | 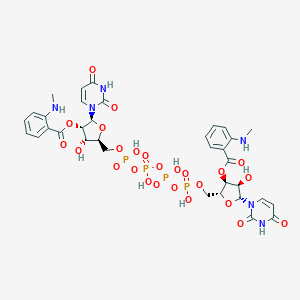 CHEMBL1162181 CHEMBL1162181 | C34H40N6O25P4 | 1056.61 | 27 / 10 | -3.6 | No |
| 90104 | 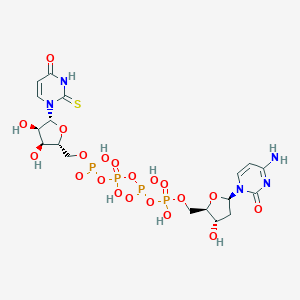 CHEMBL509434 CHEMBL509434 | C18H27N5O20P4S | 789.384 | 21 / 9 | -7.8 | No |
| 90257 |  CHEMBL1083262 CHEMBL1083262 | C23H31N2O21P3 | 764.415 | 21 / 9 | -6.1 | No |
| 91000 |  PSB-10211 PSB-10211 | C23H13Cl2N6NaO5S | 579.344 | 11 / 3 | N/A | No |
| 91125 |  CHEMBL505403 CHEMBL505403 | C19H27N7O21P4 | 813.348 | 23 / 10 | -8.6 | No |
| 91695 |  CHEMBL1199751 CHEMBL1199751 | C16H20N2O11P2S | 510.347 | 12 / 5 | -1.5 | No |
| 94100 | 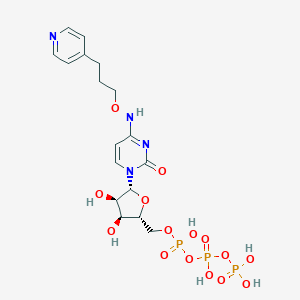 CHEMBL3261371 CHEMBL3261371 | C17H25N4O15P3 | 618.321 | 16 / 7 | -4.8 | No |
| 97496 | 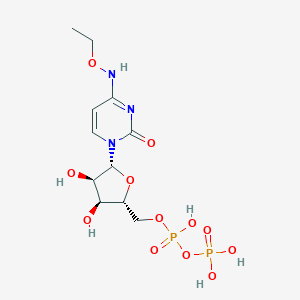 CHEMBL1084295 CHEMBL1084295 | C11H19N3O12P2 | 447.23 | 12 / 6 | -3.7 | No |
| 100342 | 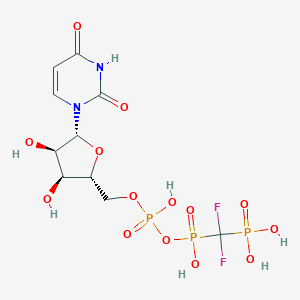 CHEMBL1765117 CHEMBL1765117 | C10H15F2N2O14P3 | 518.148 | 16 / 7 | -5.2 | No |
| 102173 | 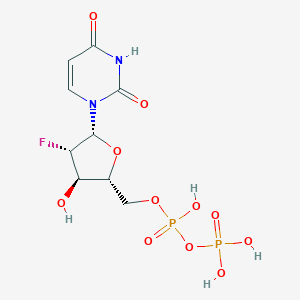 CHEMBL1199752 CHEMBL1199752 | C9H13FN2O11P2 | 406.152 | 12 / 5 | -3.7 | No |
| 102460 | 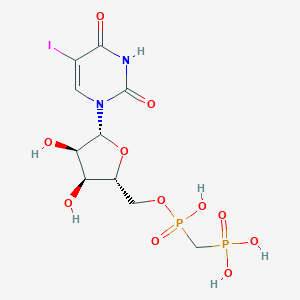 CHEMBL1085561 CHEMBL1085561 | C10H15IN2O11P2 | 528.085 | 11 / 6 | -4.5 | No |
| 102531 | 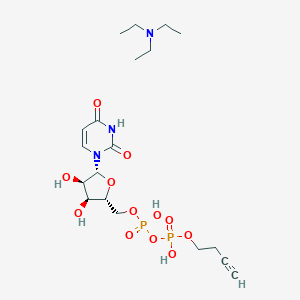 CHEMBL612064 CHEMBL612064 | C19H33N3O12P2 | 557.43 | 13 / 5 | N/A | No |
| 104007 | 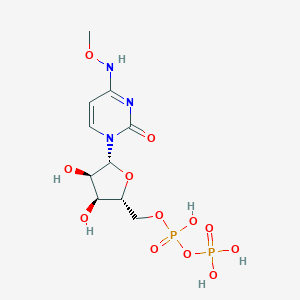 CHEMBL1084020 CHEMBL1084020 | C10H17N3O12P2 | 433.203 | 12 / 6 | -4.1 | No |
| 109441 | 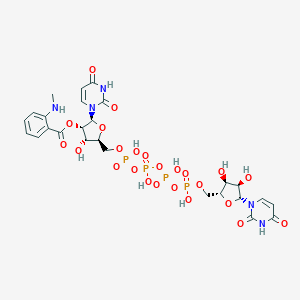 CHEMBL1162186 CHEMBL1162186 | C26H33N5O24P4 | 923.456 | 25 / 10 | -6.3 | No |
| 109710 | 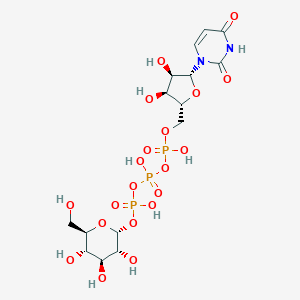 CHEMBL1083260 CHEMBL1083260 | C15H25N2O20P3 | 646.28 | 20 / 10 | -7.4 | No |
| 109886 | 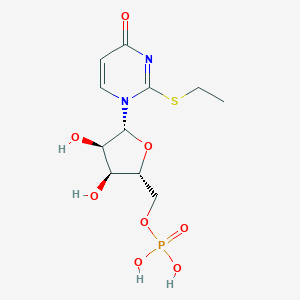 CHEMBL1767418 CHEMBL1767418 | C11H17N2O8PS | 368.297 | 9 / 4 | -1.5 | Yes |
| 110375 | 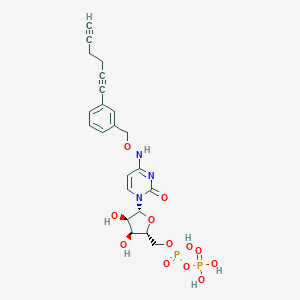 CHEMBL3220051 CHEMBL3220051 | C22H25N3O12P2 | 585.399 | 12 / 6 | -2.2 | No |
| 110821 | 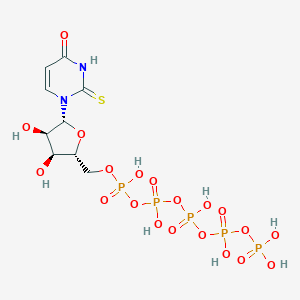 CHEMBL482683 CHEMBL482683 | C9H17N2O20P5S | 660.158 | 21 / 9 | -7.4 | No |
| 110827 | 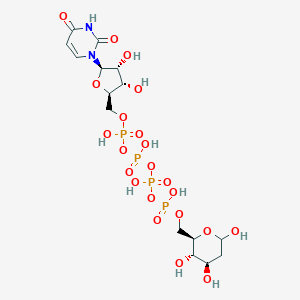 CHEMBL454230 CHEMBL454230 | C15H26N2O22P4 | 710.26 | 22 / 10 | -8.1 | No |
| 113311 | 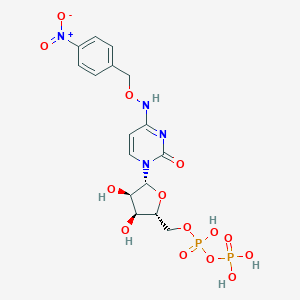 CHEMBL3220050 CHEMBL3220050 | C16H20N4O14P2 | 554.298 | 14 / 6 | -3.6 | No |
| 113654 | 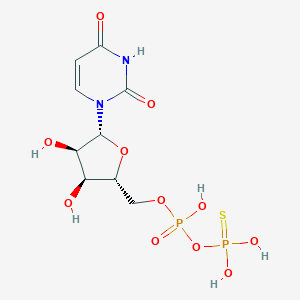 UDP-beta-S UDP-beta-S | C9H14N2O11P2S | 420.222 | 12 / 6 | -3.0 | No |
| 115803 | 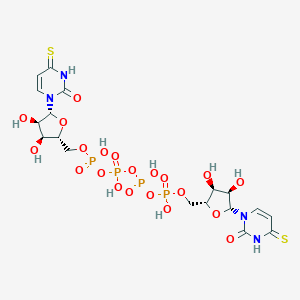 CHEMBL503798 CHEMBL503798 | C18H26N4O21P4S2 | 822.428 | 23 / 10 | -7.9 | No |
| 120596 |  CHEMBL448985 CHEMBL448985 | C14H24N2O22P4 | 696.233 | 22 / 10 | -8.4 | No |
| 122640 |  UDP-glucose UDP-glucose | C15H24N2O17P2 | 566.302 | 17 / 9 | -6.3 | No |
| 122941 |  CHEMBL3261370 CHEMBL3261370 | C17H25N4O15P3 | 618.321 | 16 / 7 | -4.8 | No |
| 124781 |  CHEMBL1161879 CHEMBL1161879 | C15H29N3O11P2S | 521.415 | 13 / 6 | N/A | No |
zhanglab![]() zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218