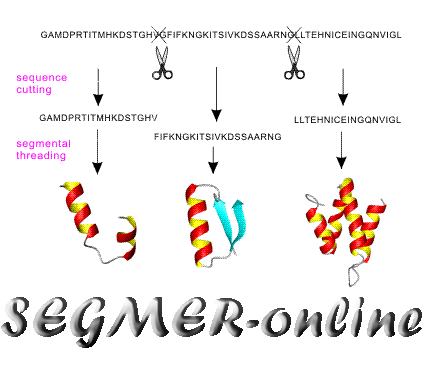
SEGMER is a segmental threading algorithm designed to recoginzing
substructure motifs from the Protein Data Bank (PDB) library.
It first splits target sequences into segments which consists of 2-4
consecutive or non-consecutive secondary structure elements (alpha-helix,
beta-strand). The sequence segments are then threaded through the PDB
to identify conserved substructures.
It often identifies better conserved structure motifs than
the whole-chain threading
methods, especially when there is no similar
global fold existing in the PDB.
The SEGMER program is downloadable at
the download page.
Reference:
S. Wu, Y. Zhang.
Recognizing Protein Substructure Similarity Using Segmental Threading.
Structure, 2010 18: 858-867
(download the PDF file).


