

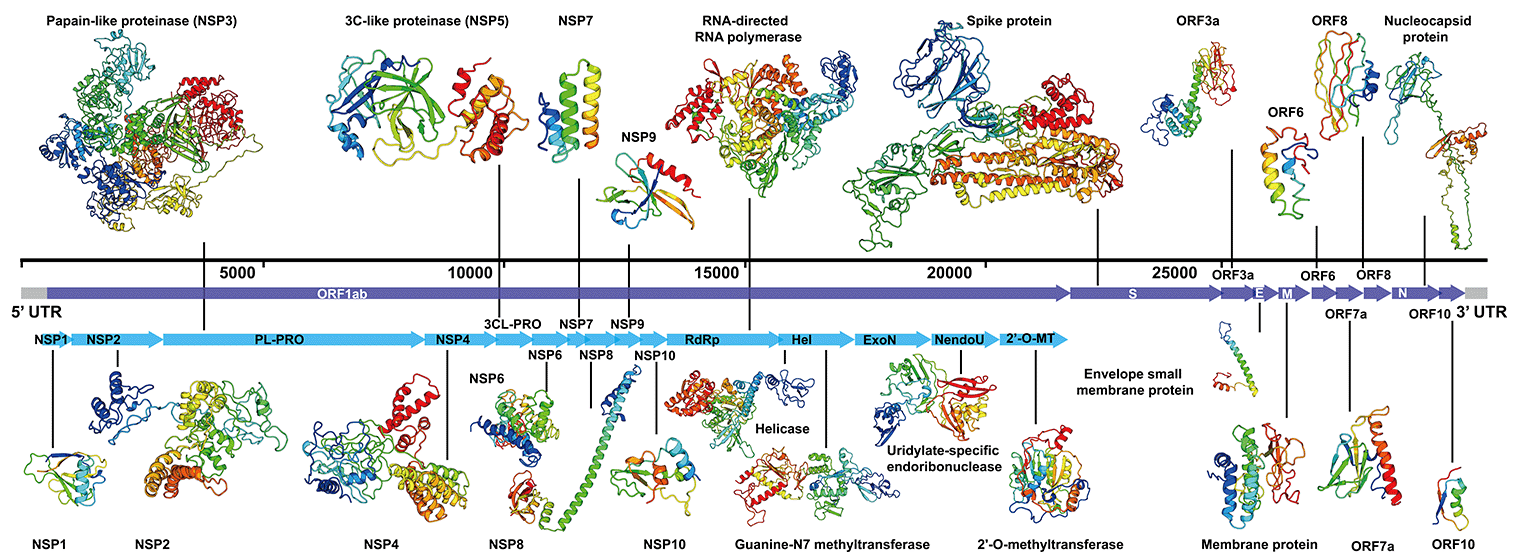

| # | Protein sequence translated from SARS-CoV-2 genome | D-I-TASSER structure model and estimated accuracy | Protein name and function (based on UniProt curation of SARS-CoV-2 proteome) | Solved experimental structure |
| 1 | >QHD43415_1
(L=180) MESLVPGFNEKTHVQLSLPVLQVRDVLVRGFGDSVEEVLS EARQHLKDGTCGLVEVEKGVLPQLEQPYVFIKRSDARTAP HGHVMVELVAELEGIQYGRSGETLGVLVPHVGEIPVAYRK VLLRKNGNKGAGGHSYGADLKSFDLGDELGTDPYEDFQEN WNTKHSSGVTRELMRELNGG |
 [QHD43415_1.pdb] Estimate TM-score=0.87 |
Host translation inhibitor nsp1. Inhibits host translation by interacting with the 40S ribosomal subunit. The nsp1-40S ribosome complex further induces an endonucleolytic cleavage near the 5'UTR of host mRNAs, targeting them for degradation. Viral mRNAs are not susceptible to nsp1-mediated endonucleolytic RNA cleavage thanks to the presence of a 5'-end leader sequence and are therefore protected from degradation. By suppressing host gene expression, nsp1 facilitates efficient viral gene expression in infected cells and evasion from host immune response. |
7K3N(range:1-180); |
| 2 | >QHD43415_2
(L=638) AYTRYVDNNFCGPDGYPLECIKDLLARAGKASCTLSEQLD FIDTKRGVYCCREHEHEIAWYTERSEKSYELQTPFEIKLA KKFDTFNGECPNFVFPLNSIIKTIQPRVEKKKLDGFMGRI RSVYPVASPNECNQMCLSTLMKCDHCGETSWQTGDFVKAT CEFCGTENLTKEGATTCGYLPQNAVVKIYCPACHNSEVGP EHSLAEYHNESGLKTILRKGGRTIAFGGCVFSYVGCHNKC AYWVPRASANIGCNHTGVVGEGSEGLNDNLLEILQKEKVN INIVGDFKLNEEIAIILASFSASTSAFVETVKGLDYKAFK QIVESCGNFKVTKGKAKKGAWNIGEQKSILSPLYAFASEA ARVVRSIFSRTLETAQNSVRVLQKAAITILDGISQYSLRL IDAMMFTSDLATNNLVVMAYITGGVVQLTSQWLTNIFGTV YEKLKPVLDWLEEKFKEGVEFLRDGWEIVKFISTCACEIV GGQIVTCAKEIKESVQTFFKLVNKFLALCADSIIIGGAKL KALNLGETFVTHSKGLYRKCVKSREETGLLMPLKAPKEII FLEGETLPTEVLTEEVVLKTGDLQPLEQPTSEAVEAPLVG TPVCINGLMLLEIKDTEKYCALAPNMMVTNNTFTLKGG |
 [QHD43415_2.pdb] Estimate TM-score=0.62 |
Non-structural protein 2 (nsp2). May play a role in the modulation of host cell survival signaling pathway by interacting with host PHB and PHB2. Indeed, these two proteins play a role in maintaining the functional integrity of the mitochondria and protecting cells from various stresses. |
NA(range:NA); |
| 3 | >QHD43415_3
(L=1945) APTKVTFGDDTVIEVQGYKSVNITFELDERIDKVLNEKCS AYTVELGTEVNEFACVVADAVIKTLQPVSELLTPLGIDLD EWSMATYYLFDESGEFKLASHMYCSFYPPDEDEEEGDCEE EEFEPSTQYEYGTEDDYQGKPLEFGATSAALQPEEEQEED WLDDDSQQTVGQQDGSEDNQTTTIQTIVEVQPQLEMELTP VVQTIEVNSFSGYLKLTDNVYIKNADIVEEAKKVKPTVVV NAANVYLKHGGGVAGALNKATNNAMQVESDDYIATNGPLK VGGSCVLSGHNLAKHCLHVVGPNVNKGEDIQLLKSAYENF NQHEVLLAPLLSAGIFGADPIHSLRVCVDTVRTNVYLAVF DKNLYDKLVSSFLEMKSEKQVEQKIAEIPKEEVKPFITES KPSVEQRKQDDKKIKACVEEVTTTLEETKFLTENLLLYID INGNLHPDSATLVSDIDITFLKKDAPYIVGDVVQEGVLTA VVIPTKKAGGTTEMLAKALRKVPTDNYITTYPGQGLNGYT VEEAKTVLKKCKSAFYILPSIISNEKQEILGTVSWNLREM LAHAEETRKLMPVCVETKAIVSTIQRKYKGIKIQEGVVDY GARFYFYTSKTTVASLINTLNDLNETLVTMPLGYVTHGLN LEEAARYMRSLKVPATVSVSSPDAVTAYNGYLTSSSKTPE EHFIETISLAGSYKDWSYSGQSTQLGIEFLKRGDKSVYYT SNPTTFHLDGEVITFDNLKTLLSLREVRTIKVFTTVDNIN LHTQVVDMSMTYGQQFGPTYLDGADVTKIKPHNSHEGKTF YVLPNDDTLRVEAFEYYHTTDPSFLGRYMSALNHTKKWKY PQVNGLTSIKWADNNCYLATALLTLQQIELKFNPPALQDA YYRARAGEAANFCALILAYCNKTVGELGDVRETMSYLFQH ANLDSCKRVLNVVCKTCGQQQTTLKGVEAVMYMGTLSYEQ FKKGVQIPCTCGKQATKYLVQQESPFVMMSAPPAQYELKH GTFTCASEYTGNYQCGHYKHITSKETLYCIDGALLTKSSE YKGPITDVFYKENSYTTTIKPVTYKLDGVVCTEIDPKLDN YYKKDNSYFTEQPIDLVPNQPYPNASFDNFKFVCDNIKFA DDLNQLTGYKKPASRELKVTFFPDLNGDVVAIDYKHYTPS FKKGAKLLHKPIVWHVNNATNKATYKPNTWCIRCLWSTKP VETSNSFDVLKSEDAQGMDNLACEDLKPVSEEVVENPTIQ KDVLECNVKTTEVVGDIILKPANNSLKITEEVGHTDLMAA YVDNSSLTIKKPNELSRVLGLKTLATHGLAAVNSVPWDTI ANYAKPFLNKVVSTTTNIVTRCLNRVCTNYMPYFFTLLLQ LCTFTRSTNSRIKASMPTTIAKNTVKSVGKFCLEASFNYL KSPNFSKLINIIIWFLLLSVCLGSLIYSTAALGVLMSNLG MPSYCTGYREGYLNSTNVTIATYCTGSIPCSVCLSGLDSL DTYPSLETIQITISSFKWDLTAFGLVAEWFLAYILFTRFF YVLGLAAIMQLFFSYFAVHFISNSWLMWLIINLVQMAPIS AMVRMYIFFASFYYVWKSYVHVVDGCNSSTCMMCYKRNRA TRVECTTIVNGVRRSFYVYANGGKGFCKLHNWNCVNCDTF CAGSTFISDEVARDLSLQFKRPINPTDQSSYIVDSVTVKN GSIHLYFDKAGQKTYERHSLSHFVNLDNLRANNTKGSLPI NVIVFDGKSKCEESSAKSASVYYSQLMCQPILLLDQALVS DVGDSAEVAVKMFDAYVNTFSSTFNVPMEKLKTLVATAEA ELAKNVSLDNVLSTFISAARQGFVDSDVETKDVVECLKLS HQSDIEVTGDSCNNYMLTYNKVENMTPRDLGACIDCSARH INAQVAKSHNIALIWNVKDFMSLSEQLRKQIRSAAKKNNL PFKLTCATTRQVVNVVTTKIALKGG |
 [QHD43415_3.pdb] Estimate TM-score=0.90 |
Papain-like proteinase. Responsible for the cleavages located at the N-terminus of the replicase polyprotein. In addition, PL-PRO possesses a deubiquitinating/deISGylating activity and processes both 'Lys-48'- and 'Lys-63'-linked polyubiquitin chains from cellular substrates. Participates together with nsp4 in the assembly of virally-induced cytoplasmic double-membrane vesicles necessary for viral replication. Antagonizes innate immune induction of type I interferon by blocking the phosphorylation, dimerization and subsequent nuclear translocation of host IRF3. Prevents also host NF-kappa-B signaling. |
7KAG(range:1-111);6W6Y(range:207-379);6W9C(range:748-1060); |
| 4 | >QHD43415_4
(L=500) KIVNNWLKQLIKVTLVFLFVAAIFYLITPVHVMSKHTDFS SEIIGYKAIDGGVTRDIASTDTCFANKHADFDTWFSQRGG SYTNDKACPLIAAVITREVGFVVPGLPGTILRTTNGDFLH FLPRVFSAVGNICYTPSKLIEYTDFATSACVLAAECTIFK DASGKPVPYCYDTNVLEGSVAYESLRPDTRYVLMDGSIIQ FPNTYLEGSVRVVTTFDSEYCRHGTCERSEAGVCVSTSGR WVLNNDYYRSLPGVFCGVDAVNLLTNMFTPLIQPIGALDI SASIVAGGIVAIVVTCLAYYFMRFRRAFGEYSHVVAFNTL LFLMSFTVLCLTPVYSFLPGVYSVIYLYLTFYLTNDVSFL AHIQWMVMFTPLVPFWITIAYIICISTKHFYWFFSNYLKR RVVFNGVSFSTFEEAALCTFLLNKEMYLKLRSDVLLPLTQ YNRYLALYNKYKYFSGAMDTTSYREAACCHLAKALNDFSN SGSDVLYQPPQTSITSAVLQ |
 [QHD43415_4.pdb] Estimate TM-score=0.61 |
Non-structural protein 4 (nsp4). Participates in the assembly of virally-induced cytoplasmic double-membrane vesicles necessary for viral replication. |
NA(range:NA); |
| 5 | >QHD43415_5
(L=306) SGFRKMAFPSGKVEGCMVQVTCGTTTLNGLWLDDVVYCPR HVICTSEDMLNPNYEDLLIRKSNHNFLVQAGNVQLRVIGH SMQNCVLKLKVDTANPKTPKYKFVRIQPGQTFSVLACYNG SPSGVYQCAMRPNFTIKGSFLNGSCGSVGFNIDYDCVSFC YMHHMELPTGVHAGTDLEGNFYGPFVDRQTAQAAGTDTTI TVNVLAWLYAAVINGDRWFLNRFTTTLNDFNLVAMKYNYE PLTQDHVDILGPLSAQTGIAVLDMCASLKELLQNGMNGRT ILGSALLEDEFTPFDVVRQCSGVTFQ |
 [QHD43415_5.pdb] Estimate TM-score=0.97 |
Proteinase 3CL-PRO. Cleaves the C-terminus of replicase polyprotein at 11 sites. Recognizes substrates containing the core sequence [ILMVF]-Q-|-[SGACN]. Also able to bind an ADP-ribose-1''-phosphate (ADRP). |
6LU7(range:1-306); |
| 6 | >QHD43415_6
(L=290) SAVKRTIKGTHHWLLLTILTSLLVLVQSTQWSLFFFLYEN AFLPFAMGIIAMSAFAMMFVKHKHAFLCLFLLPSLATVAY FNMVYMPASWVMRIMTWLDMVDTSLSGFKLKDCVMYASAV VLLILMTARTVYDDGARRVWTLMNVLTLVYKVYYGNALDQ AISMWALIISVTSNYSGVVTTVMFLARGIVFMCVEYCPIF FITGNTLQCIMLVYCFLGYFCTCYFGLFCLLNRYFRLTLG VYDYLVSTQEFRYMNSQGLLPPKNSIDAFKLNIKLLGVGG KPCIKVATVQ |
 [QHD43415_6.pdb] Estimate TM-score=0.52 |
Non-structural protein 6 (nsp6). Plays a role in the initial induction of autophagosomes from host reticulum endoplasmic. Later, limits the expansion of these phagosomes that are no longer able to deliver viral components to lysosomes. |
NA(range:NA); |
| 7 | >QHD43415_7
(L=83) SKMSDVKCTSVVLLSVLQQLRVESSSKLWAQCVQLHNDIL LAKDTTEAFEKMVSLLSVLLSMQGAVDINKLCEEMLDNRA TLQ |
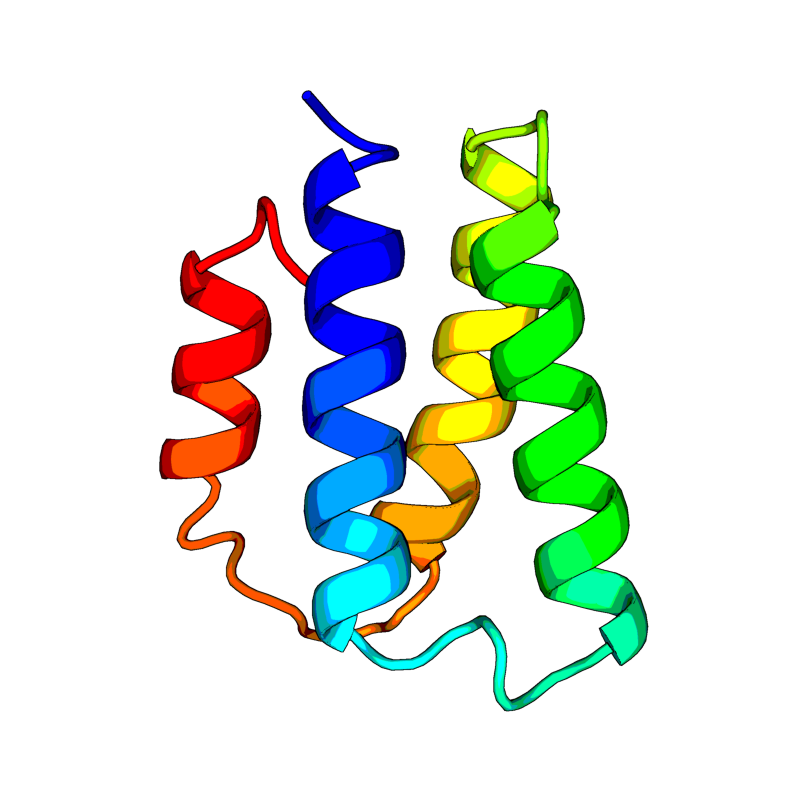 [QHD43415_7.pdb] Estimate TM-score=0.92 |
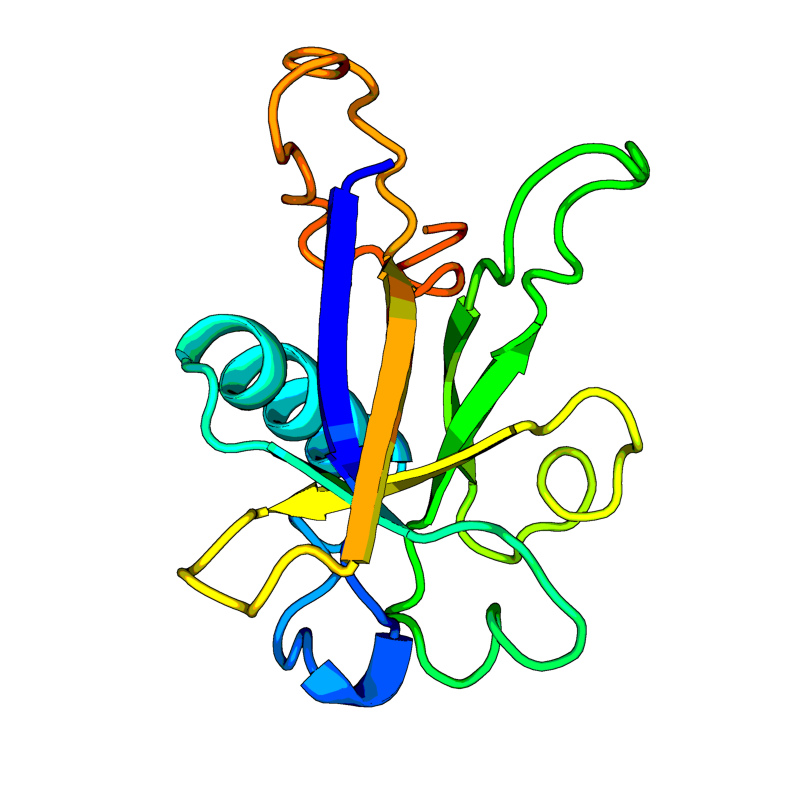
Non-structural protein 7 (nsp7). Forms a hexadecamer with nsp8 (8 subunits of each) that may participate in viral replication by acting as a primase. Alternatively, may synthesize substantially longer products than oligonucleotide primers. |
6M71(range:1-83); |
| 8 | >QHD43415_8
(L=198) AIASEFSSLPSYAAFATAQEAYEQAVANGDSEVVLKKLKK SLNVAKSEFDRDAAMQRKLEKMADQAMTQMYKQARSEDKR AKVTSAMQTMLFTMLRKLDNDALNNIINNARDGCVPLNII PLTTAAKLMVVIPDYNTYKNTCDGTTFTYASALWEIQQVV DADSKIVQLSEISMDNSPNLAWPLIVTALRANSAVKLQ |
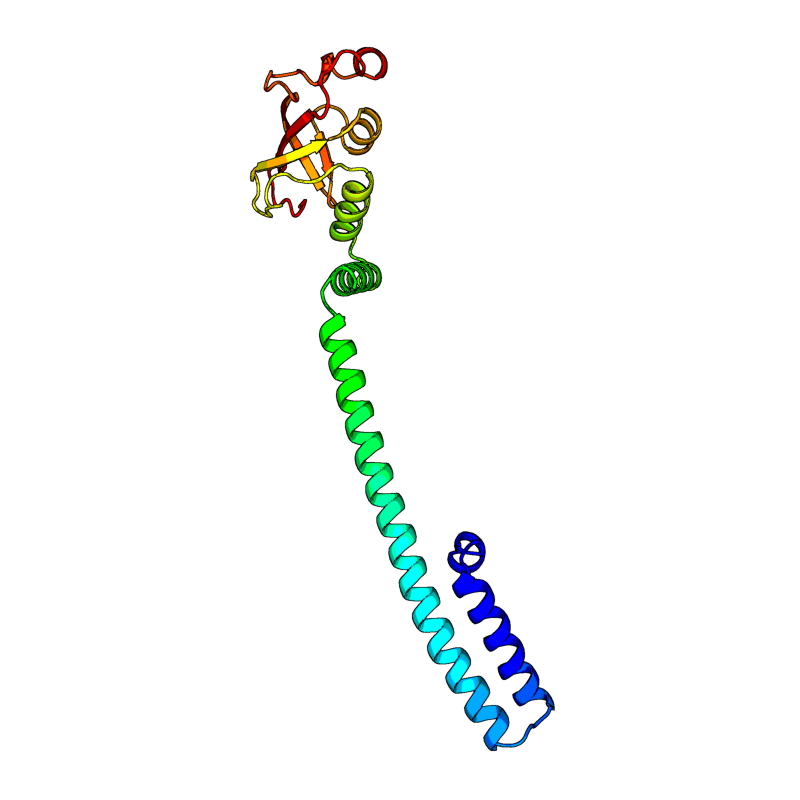 [QHD43415_8.pdb] Estimate TM-score=0.95 |
Non-structural protein 8 (nsp8). Forms a hexadecamer with nsp7 (8 subunits of each) that may participate in viral replication by acting as a primase. Alternatively, may synthesize substantially longer products than oligonucleotide primers. |
7CYQ(range:1-198); |
| 9 | >QHD43415_9
(L=113) NNELSPVALRQMSCAAGTTQTACTDDNALAYYNTTKGGRF VLALLSDLQDLKWARFPKSDGTGTIYTELEPPCRFVTDTP KGPKVKYLYFIKGLNNLNRGMVLGSLAATVRLQ |
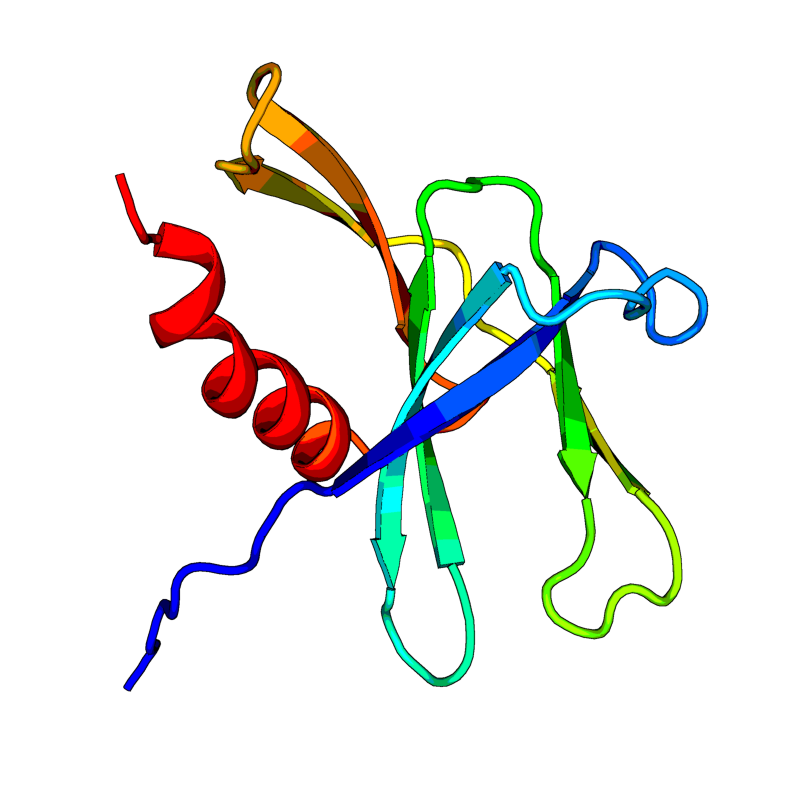 [QHD43415_9.pdb] Estimate TM-score=0.91 |
Non-structural protein 9 (nsp9). May participate in viral replication by acting as a ssRNA-binding protein. |
6W4B(range:1-113); |
| 10 | >QHD43415_10
(L=139) AGNATEVPANSTVLSFCAFAVDAAKAYKDYLASGGQPITN CVKMLCTHTGTGQAITVTPEANMDQESFGGASCCLYCRCH IDHPNPKGFCDLKGKYVQIPTTCANDPVGFTLKNTVCTVC GMWKGYGCSCDQLREPMLQ |
 [QHD43415_10.pdb] Estimate TM-score=0.90 |
Non-structural protein 10 (nsp10). Plays a pivotal role in viral transcription by stimulating both nsp14 3'-5' exoribonuclease and nsp16 2'-O-methyltransferase activities. Therefore plays an essential role in viral mRNAs cap methylation. |
6W75(range:1-139); |
| 11 | >QHD43415_11
(L=932) SADAQSFLNRVCGVSAARLTPCGTGTSTDVVYRAFDIYND KVAGFAKFLKTNCCRFQEKDEDDNLIDSYFVVKRHTFSNY QHEETIYNLLKDCPAVAKHDFFKFRIDGDMVPHISRQRLT KYTMADLVYALRHFDEGNCDTLKEILVTYNCCDDDYFNKK DWYDFVENPDILRVYANLGERVRQALLKTVQFCDAMRNAG IVGVLTLDNQDLNGNWYDFGDFIQTTPGSGVPVVDSYYSL LMPILTLTRALTAESHVDTDLTKPYIKWDLLKYDFTEERL KLFDRYFKYWDQTYHPNCVNCLDDRCILHCANFNVLFSTV FPPTSFGPLVRKIFVDGVPFVVSTGYHFRELGVVHNQDVN LHSSRLSFKELLVYAADPAMHAASGNLLLDKRTTCFSVAA LTNNVAFQTVKPGNFNKDFYDFAVSKGFFKEGSSVELKHF FFAQDGNAAISDYDYYRYNLPTMCDIRQLLFVVEVVDKYF DCYDGGCINANQVIVNNLDKSAGFPFNKWGKARLYYDSMS YEDQDALFAYTKRNVIPTITQMNLKYAISAKNRARTVAGV SICSTMTNRQFHQKLLKSIAATRGATVVIGTSKFYGGWHN MLKTVYSDVENPHLMGWDYPKCDRAMPNMLRIMASLVLAR KHTTCCSLSHRFYRLANECAQVLSEMVMCGGSLYVKPGGT SSGDATTAYANSVFNICQAVTANVNALLSTDGNKIADKYV RNLQHRLYECLYRNRDVDTDFVNEFYAYLRKHFSMMILSD DAVVCFNSTYASQGLVASIKNFKSVLYYQNNVFMSEAKCW TETDLTKGPHEFCSQHTMLVKQGDDYVYLPYPDPSRILGA GCFVDDIVKTDGTLMIERFVSLAIDAYPLTKHPNQEYADV FHLYLQYIRKLHDELTGHMLDMYSVMLTNDNTSRYWEPEF YEAMYTPHTVLQ |
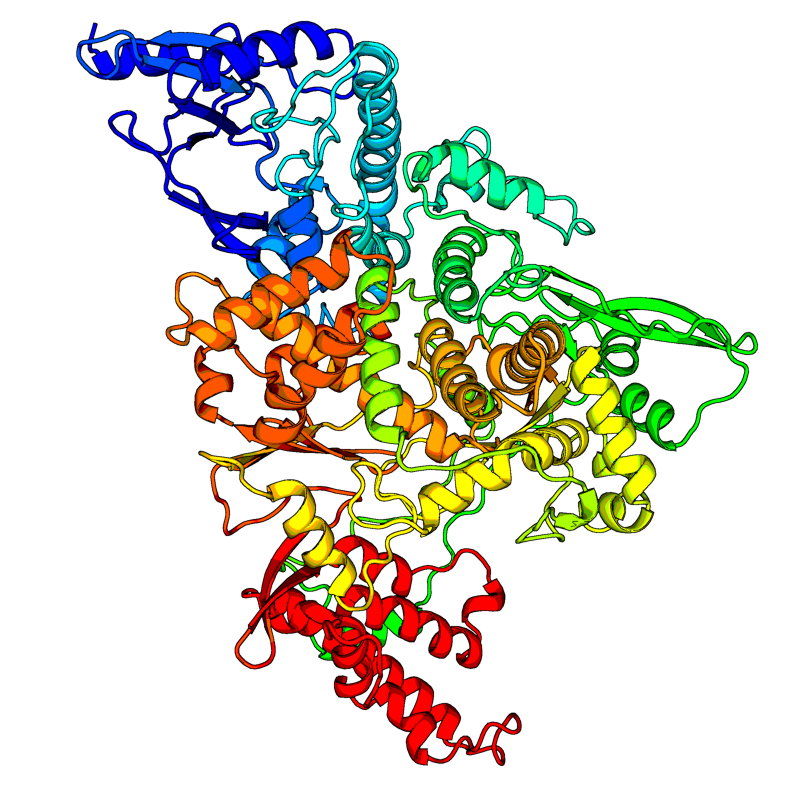 [QHD43415_11.pdb] Estimate TM-score=0.99 |
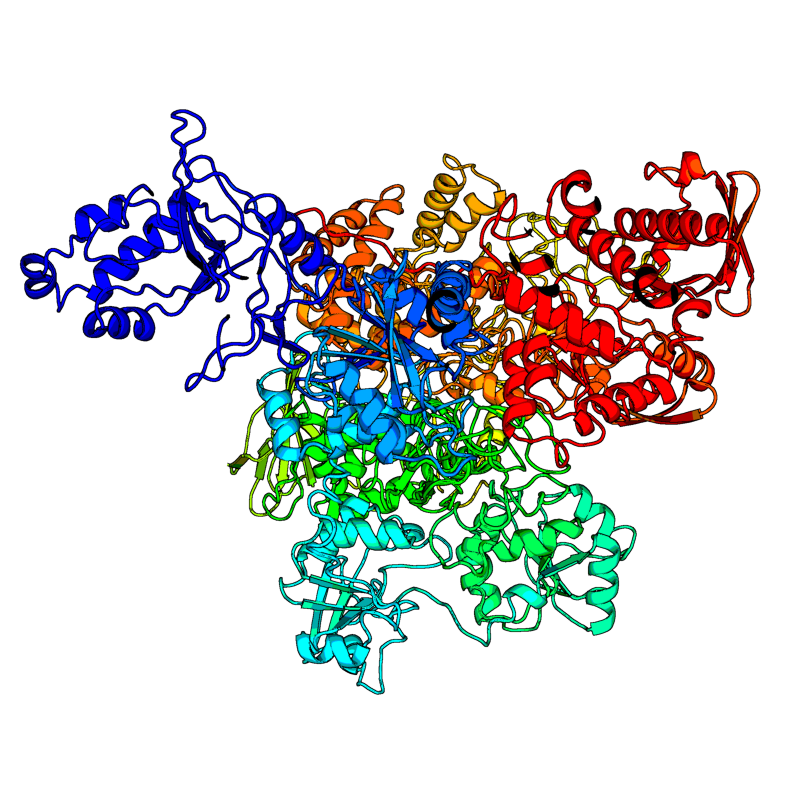
RNA-directed RNA polymerase (RdRp). Responsible for replication and transcription of the viral RNA genome. |
6M71(range:1-932); |
| 12 | >QHD43415_12
(L=601) AVGACVLCNSQTSLRCGACIRRPFLCCKCCYDHVISTSHK LVLSVNPYVCNAPGCDVTDVTQLYLGGMSYYCKSHKPPIS FPLCANGQVFGLYKNTCVGSDNVTDFNAIATCDWTNAGDY ILANTCTERLKLFAAETLKATEETFKLSYGIATVREVLSD RELHLSWEVGKPRPPLNRNYVFTGYRVTKNSKVQIGEYTF EKGDYGDAVVYRGTTTYKLNVGDYFVLTSHTVMPLSAPTL VPQEHYVRITGLYPTLNISDEFSSNVANYQKVGMQKYSTL QGPPGTGKSHFAIGLALYYPSARIVYTACSHAAVDALCEK ALKYLPIDKCSRIIPARARVECFDKFKVNSTLEQYVFCTV NALPETTADIVVFDEISMATNYDLSVVNARLRAKHYVYIG DPAQLPAPRTLLTKGTLEPEYFNSVCRLMKTIGPDMFLGT CRRCPAEIVDTVSALVYDNKLKAHKDKSAQCFKMFYKGVI THDVSSAINRPQIGVVREFLTRNPAWRKAVFISPYNSQNA VASKILGLPTQTVDSSQGSEYDYVIFTQTTETAHSCNVNR FNVAITRAKVGILCIMSDRDLYDKLQFTSLEIPRRNVATL Q |
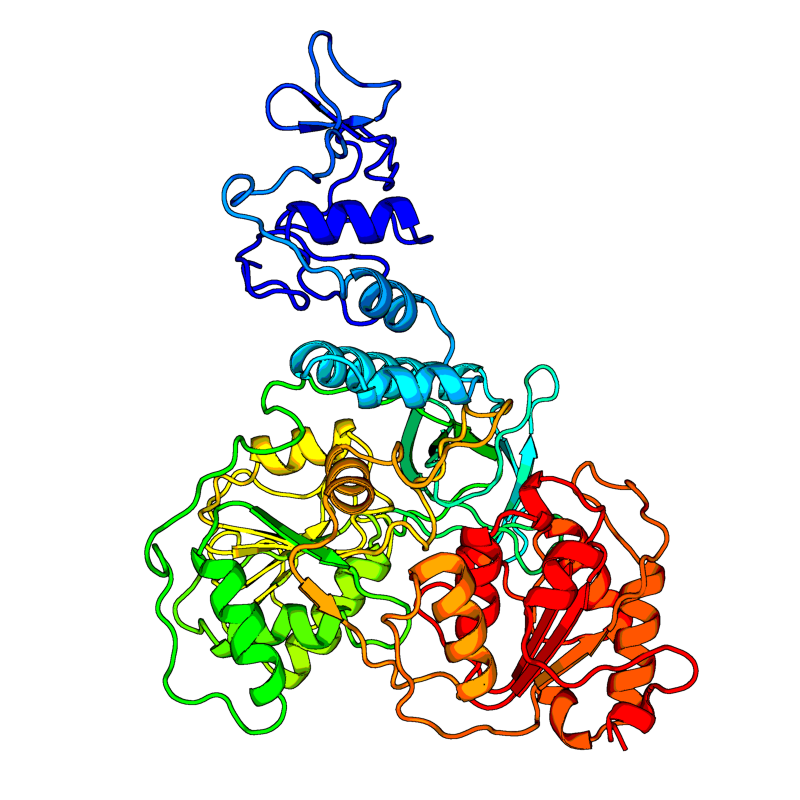 [QHD43415_12.pdb] Estimate TM-score=0.95 |
Helicase (Hel). Multi-functional protein with a zinc-binding domain in N-terminus displaying RNA and DNA duplex-unwinding activities with 5' to 3' polarity. Activity of helicase is dependent on magnesium. |
5RL9(range:1-601); |
| 13 | >QHD43415_13
(L=527) AENVTGLFKDCSKVITGLHPTQAPTHLSVDTKFKTEGLCV DIPGIPKDMTYRRLISMMGFKMNYQVNGYPNMFITREEAI RHVRAWIGFDVEGCHATREAVGTNLPLQLGFSTGVNLVAV PTGYVDTPNNTDFSRVSAKPPPGDQFKHLIPLMYKGLPWN VVRIKIVQMLSDTLKNLSDRVVFVLWAHGFELTSMKYFVK IGPERTCCLCDRRATCFSTASDTYACWHHSIGFDYVYNPF MIDVQQWGFTGNLQSNHDLYCQVHGNAHVASCDAIMTRCL AVHECFVKRVDWTIEYPIIGDELKINAACRKVQHMVVKAA LLADKFPVLHDIGNPKAIKCVPQADVEWKFYDAQPCSDKA YKIEELFYSYATHSDKFTDGVCLFWNCNVDRYPANSIVCR FDTRVLSNLNLPGCDGGSLYVNKHAFHTPAFDKSAFVNLK QLPFFYYSDSPCESHGKQVVSDIDYVPLKSATCITRCNLG GAVCRHHANEYRLYLDAYNMMISAGFSLWVYKQFDTYNLW NTFTRLQ |
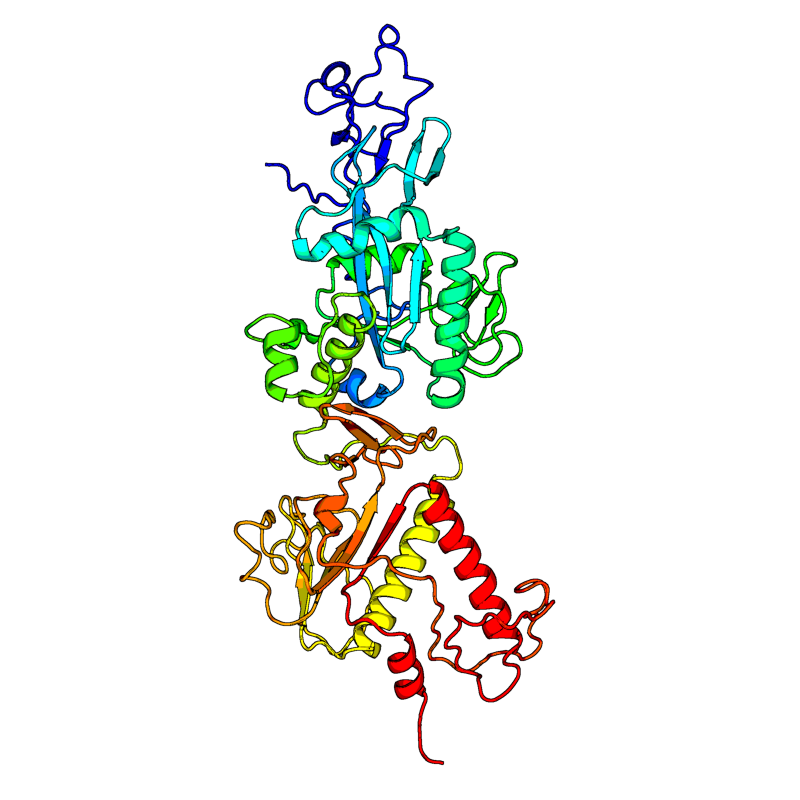 [QHD43415_13.pdb] Estimate TM-score=0.87 |
Proofreading exoribonuclease / Guanine-N7 methyltransferase (ExoN). Enzyme possessing two different activities: an exoribonuclease activity acting on both ssRNA and dsRNA in a 3' to 5' direction and a N7-guanine methyltransferase activity. Acts as a proofreading exoribonuclease for RNA replication, thereby lowering the sensitivity of the virus to RNA mutagens. |
NA(range:NA); |
| 14 | >QHD43415_14
(L=346) SLENVAFNVVNKGHFDGQQGEVPVSIINNTVYTKVDGVDV ELFENKTTLPVNVAFELWAKRNIKPVPEVKILNNLGVDIA ANTVIWDYKRDAPAHISTIGVCSMTDIAKKPTETICAPLT VFFDGRVDGQVDLFRNARNGVLITEGSVKGLQPSVGPKQA SLNGVTLIGEAVKTQFNYYKKVDGVVQQLPETYFTQSRNL QEFKPRSQMEIDFLELAMDEFIERYKLEGYAFEHIVYGDF SHSQLGGLHLLIGLAKRFKESPFELEDFIPMDSTVKNYFI TDAQTGSSKCVCSVIDLLLDDFVEIIKSQDLSVVSKVVKV TIDYTEISFMLWCKDGHVETFYPKLQ |
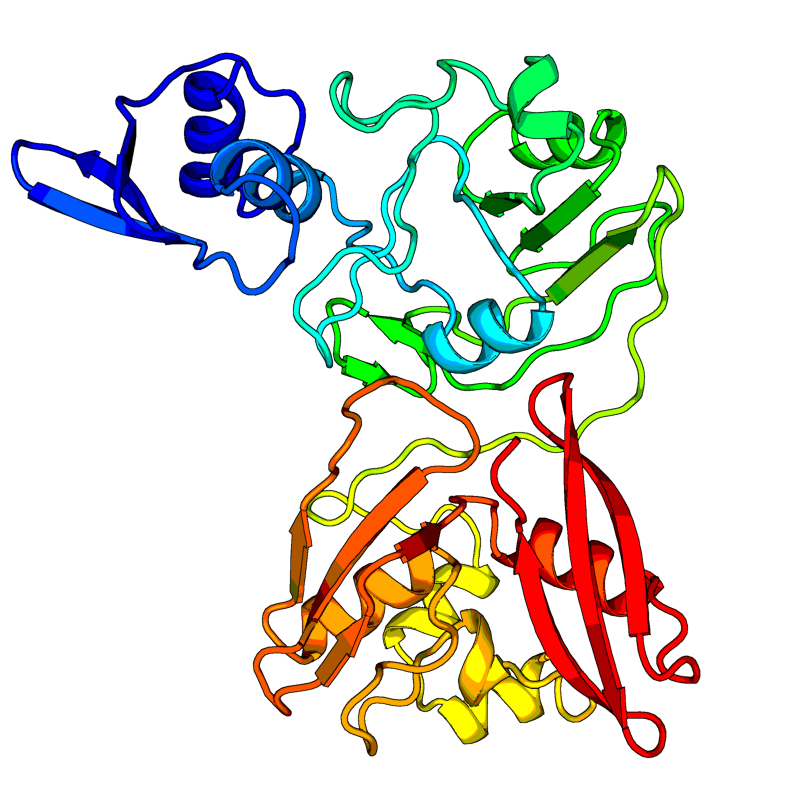 [QHD43415_14.pdb] Estimate TM-score=0.99 |
Uridylate-specific endoribonuclease (NendoU). Mn(2+)-dependent, uridylate-specific enzyme, which leaves 2'-3'-cyclic phosphates 5' to the cleaved bond. |
6VWW(range:1-346); |
| 15 | >QHD43415_15
(L=298) SSQAWQPGVAMPNLYKMQRMLLEKCDLQNYGDSATLPKGI MMNVAKYTQLCQYLNTLTLAVPYNMRVIHFGAGSDKGVAP GTAVLRQWLPTGTLLVDSDLNDFVSDADSTLIGDCATVHT ANKWDLIISDMYDPKTKNVTKENDSKEGFFTYICGFIQQK LALGGSVAIKITEHSWNADLYKLMGHFAWWTAFVTNVNAS SSEAFLIGCNYLGKPREQIDGYVMHANYIFWRNTNPIQLS SYSLFDMSKFPLKLRGTAVMSLKEGQINDMILSLLSKGRL IIRENNRVVISSDVLVNN |
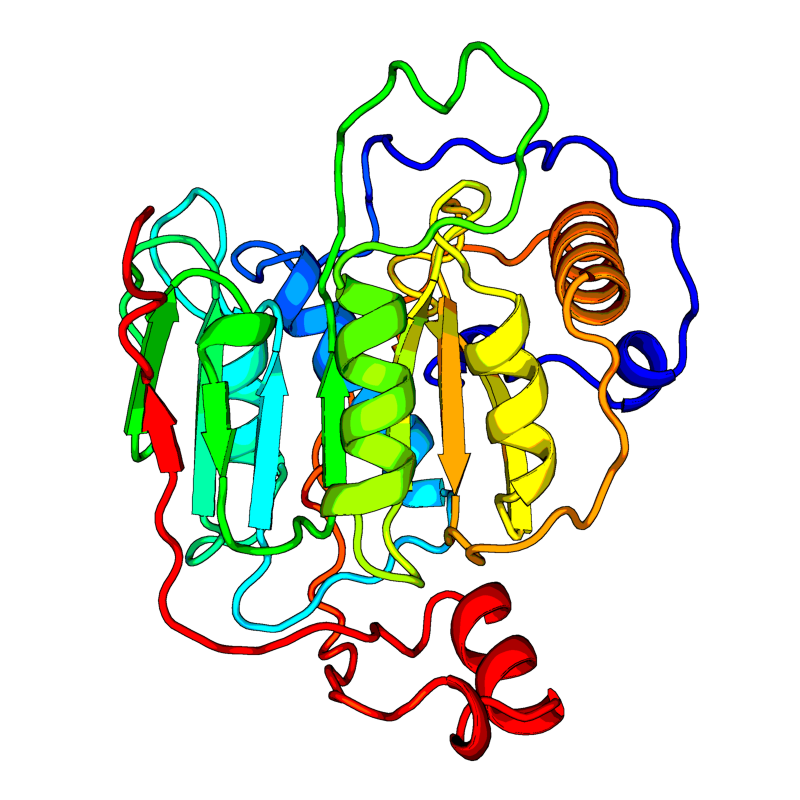 [QHD43415_15.pdb] Estimate TM-score=0.97 |
2'-O-methyltransferase (2'-O-MT). Methyltransferase that mediates mRNA cap 2'-O-ribose methylation to the 5'-cap structure of viral mRNAs. N7-methyl guanosine cap is a prerequisite for binding of nsp16. Therefore plays an essential role in viral mRNAs cap methylation which is essential to evade immune system. |
6W75(range:1-298); |
| 16 | >QHD43416
(L=1273) MFVFLVLLPLVSSQCVNLTTRTQLPPAYTNSFTRGVYYPD KVFRSSVLHSTQDLFLPFFSNVTWFHAIHVSGTNGTKRFD NPVLPFNDGVYFASTEKSNIIRGWIFGTTLDSKTQSLLIV NNATNVVIKVCEFQFCNDPFLGVYYHKNNKSWMESEFRVY SSANNCTFEYVSQPFLMDLEGKQGNFKNLREFVFKNIDGY FKIYSKHTPINLVRDLPQGFSALEPLVDLPIGINITRFQT LLALHRSYLTPGDSSSGWTAGAAAYYVGYLQPRTFLLKYN ENGTITDAVDCALDPLSETKCTLKSFTVEKGIYQTSNFRV QPTESIVRFPNITNLCPFGEVFNATRFASVYAWNRKRISN CVADYSVLYNSASFSTFKCYGVSPTKLNDLCFTNVYADSF VIRGDEVRQIAPGQTGKIADYNYKLPDDFTGCVIAWNSNN LDSKVGGNYNYLYRLFRKSNLKPFERDISTEIYQAGSTPC NGVEGFNCYFPLQSYGFQPTNGVGYQPYRVVVLSFELLHA PATVCGPKKSTNLVKNKCVNFNFNGLTGTGVLTESNKKFL PFQQFGRDIADTTDAVRDPQTLEILDITPCSFGGVSVITP GTNTSNQVAVLYQDVNCTEVPVAIHADQLTPTWRVYSTGS NVFQTRAGCLIGAEHVNNSYECDIPIGAGICASYQTQTNS PRRARSVASQSIIAYTMSLGAENSVAYSNNSIAIPTNFTI SVTTEILPVSMTKTSVDCTMYICGDSTECSNLLLQYGSFC TQLNRALTGIAVEQDKNTQEVFAQVKQIYKTPPIKDFGGF NFSQILPDPSKPSKRSFIEDLLFNKVTLADAGFIKQYGDC LGDIAARDLICAQKFNGLTVLPPLLTDEMIAQYTSALLAG TITSGWTFGAGAALQIPFAMQMAYRFNGIGVTQNVLYENQ KLIANQFNSAIGKIQDSLSSTASALGKLQDVVNQNAQALN TLVKQLSSNFGAISSVLNDILSRLDKVEAEVQIDRLITGR LQSLQTYVTQQLIRAAEIRASANLAATKMSECVLGQSKRV DFCGKGYHLMSFPQSAPHGVVFLHVTYVPAQEKNFTTAPA ICHDGKAHFPREGVFVSNGTHWFVTQRNFYEPQIITTDNT FVSGNCDVVIGIVNNTVYDPLQPELDSFKEELDKYFKNHT SPDVDLGDISGINASVVNIQKEIDRLNEVAKNLNESLIDL QELGKYEQYIKWPWYIWLGFIAGLIAIVMVTIMLCCMTSC CSCLKGCCSCGSCCKFDEDDSEPVLKGVKLHYT |
 [QHD43416.pdb] Estimate TM-score=0.98 |
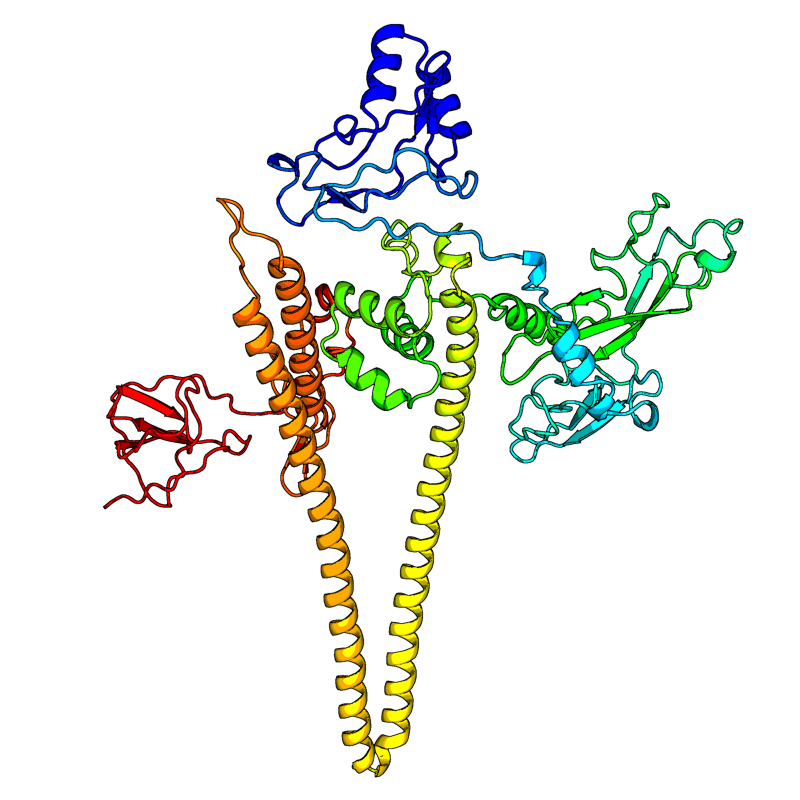
Spike glycoprotein (S). Spike protein S1 (residue 13-685): attaches the virion to the cell membrane by interacting with host receptor, initiating the infection. Binding to human ACE2 receptor and internalization of the virus into the endosomes of the host cell induces conformational changes in the Spike glycoprotein (PubMed:32142651, PubMed:32075877, PubMed:32155444). Uses also human TMPRSS2 for priming in human lung cells which is an essential step for viral entry (PubMed:32142651). Proteolysis by cathepsin CTSL may unmask the fusion peptide of S2 and activate membranes fusion within endosomes. Spike protein S2 (residue 686-1273): mediates fusion of the virion and cellular membranes by acting as a class I viral fusion protein. Under the current model, the protein has at least three conformational states: pre-fusion native state, pre-hairpin intermediate state, and post-fusion hairpin state. During viral and target cell membrane fusion, the coiled coil regions (heptad repeats) assume a trimer-of-hairpins structure, positioning the fusion peptide in close proximity to the C-terminal region of the ectodomain. The formation of this structure appears to drive apposition and subsequent fusion of viral and target cell membranes. Spike protein S2' (residue 816-1273): acts as a viral fusion peptide which is unmasked following S2 cleavage occurring upon virus endocytosis. |
6VYB (open state)(range:1-1273);6VXX (closed state)(range:1-1273);6LXT(range:912-988,1164-1202); |
| 17 | >QHD43417
(L=275) MDLFMRIFTIGTVTLKQGEIKDATPSDFVRATATIPIQAS LPFGWLIVGVALLAVFQSASKIITLKKRWQLALSKGVHFV CNLLLLFVTVYSHLLLVAAGLEAPFLYLYALVYFLQSINF VRIIMRLWLCWKCRSKNPLLYDANYFLCWHTNCYDYCIPY NSVTSSIVITSGDGTTSPISEHDYQIGGYTEKWESGVKDC VVLHSYFTSDYYQLYSTQLSTDTGVEHVTFFIYNKIVDEP EEHVQIHTIDGSSGVVNPVMEPIYDEPTTTTSVPL |
 [QHD43417.pdb] Estimate TM-score=0.98 |
ORF3a. Forms homotetrameric potassium sensitive ion channels (viroporin) and may modulate virus release. Up-regulates expression of fibrinogen subunits FGA, FGB and FGG in host lung epithelial cells. Induces apoptosis in cell culture. Downregulates the type 1 interferon receptor by inducing serine phosphorylation within the IFN alpha-receptor subunit 1 (IFNAR1) degradation motif and increasing IFNAR1 ubiquitination |
6XDC(range:1-275); |
| 18 | >QHD43418
(L=75) MYSFVSEETGTLIVNSVLLFLAFVVFLLVTLAILTALRLC AYCCNIVNVSLVKPSFYVYSRVKNLNSSRVPDLLV |
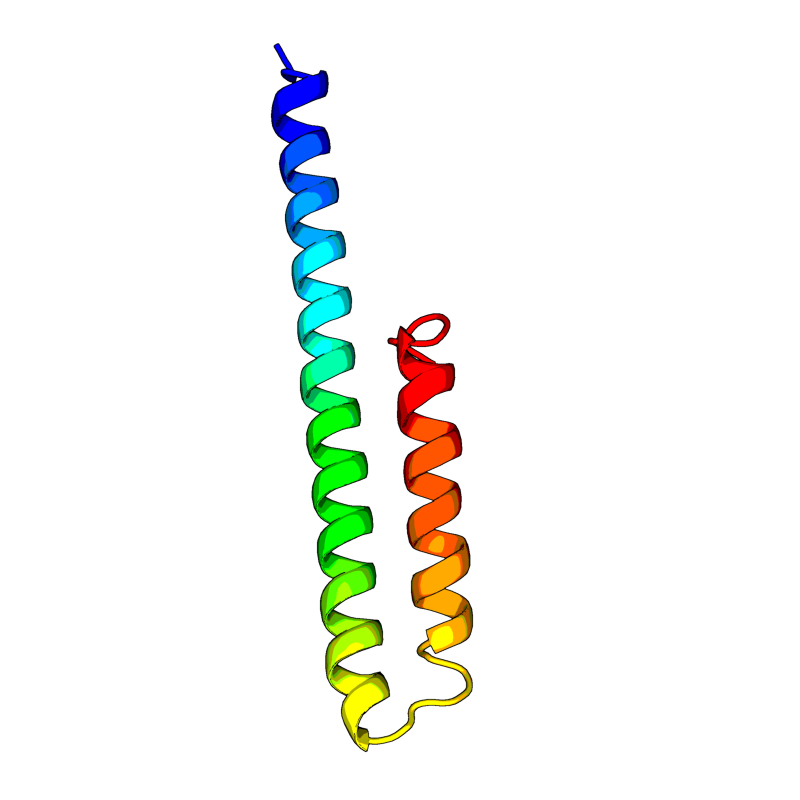 [QHD43418.pdb] Estimate TM-score=0.60 |
E. Plays a central role in virus morphogenesis and assembly. Acts as a viroporin and self-assembles in host membranes forming pentameric protein-lipid pores that allow ion transport. Also plays a role in the induction of apoptosis. Activates the host NLRP3 inflammasome, leading to IL-1beta overproduction. |
7K3G(range:8-39); |
| 19 | >QHD43419
(L=222) MADSNGTITVEELKKLLEQWNLVIGFLFLTWICLLQFAYA NRNRFLYIIKLIFLWLLWPVTLACFVLAAVYRINWITGGI AIAMACLVGLMWLSYFIASFRLFARTRSMWSFNPETNILL NVPLHGTILTRPLLESELVIGAVILRGHLRIAGHHLGRCD IKDLPKEITVATSRTLSYYKLGASQRVAGDSGFAAYSRYR IGNYKLNTDHSSSSDNIALLVQ |
 [QHD43419.pdb] Estimate TM-score=0.59 |
M. Component of the viral envelope that plays a central role in virus morphogenesis and assembly via its interactions with other viral proteins. |
NA(range:NA); |
| 20 | >QHD43420
(L=61) MFHLVDFQVTIAEILLIIMRTFKVSIWNLDYIINLIIKNL SKSLTENKYSQLDEEQPMEID |
 [QHD43420.pdb] Estimate TM-score=0.50 |
ORF6. Disrupts cell nuclear import complex formation by tethering karyopherin alpha 2 and karyopherin beta 1 to the membrane. Retention of import factors at the ER/Golgi membrane leads to a loss of transport into the nucleus. Thereby prevents STAT1 nuclear translocation in response to interferon signaling, thus blocking the expression of interferon stimulated genes (ISGs) that display multiple antiviral activities. |
NA(range:NA); |
| 21 | >QHD43421
(L=121) MKIILFLALITLATCELYHYQECVRGTTVLLKEPCSSGTY EGNSPFHPLADNKFALTCFSTQFAFACPDGVKHVYQLRAR SVSPKLFIRQEEVQELYSPIFLIVAAIVFITLCFTLKRKT E |
 [QHD43421.pdb] Estimate TM-score=0.96 |
ORF7a. Non-structural protein which is dispensable for virus replication in cell culture. |
6W37(range:16-82); |
| 22 | >QHD43422
(L=121) MKFLVFLGIITTVAAFHQECSLQSCTQHQPYVVDDPCPIH FYSKWYIRVGARKSAPLIELCVDEAGSKSPIQYIDIGNYT VSCLPFTINCQEPKLGSLVVRCSFYEDFLEYHDVRVVLDF I |
 [QHD43422.pdb] Estimate TM-score=0.99 |
ORF8. May play a role in host-virus interaction. Similar to some Bat coronavirus ns8 genes, but is entirely different from SARS ns8a or ns8b. |
7JTL(range:1-121); |
| 23 | >QHD43423
(L=419) MSDNGPQNQRNAPRITFGGPSDSTGSNQNGERSGARSKQR RPQGLPNNTASWFTALTQHGKEDLKFPRGQGVPINTNSSP DDQIGYYRRATRRIRGGDGKMKDLSPRWYFYYLGTGPEAG LPYGANKDGIIWVATEGALNTPKDHIGTRNPANNAAIVLQ LPQGTTLPKGFYAEGSRGGSQASSRSSSRSRNSSRNSTPG SSRGTSPARMAGNGGDAALALLLLDRLNQLESKMSGKGQQ QQGQTVTKKSAAEASKKPRQKRTATKAYNVTQAFGRRGPE QTQGNFGDQELIRQGTDYKHWPQIAQFAPSASAFFGMSRI GMEVTPSGTWLTYTGAIKLDDKDPNFKDQVILLNKHIDAY KTFPPTEPKKDKKKKADETQALPQRQKKQQTVTLLPAADL DDFSKQLQQSMSSADSTQA |
 [QHD43423.pdb] Estimate TM-score=0.97 |
N. Packages the positive strand viral genome RNA into a helical ribonucleocapsid (RNP) and plays a fundamental role during virion assembly through its interactions with the viral genome and membrane protein M. Plays an important role in enhancing the efficiency of subgenomic viral RNA transcription as well as viral replication. |
6M3M(range:50-174);6YUN(range:249-364); |
| 24 | >QHI42199
(L=38) MGYINVFAFPFTIYSLLLCRMNSRNYIAQVDVVNFNLT |
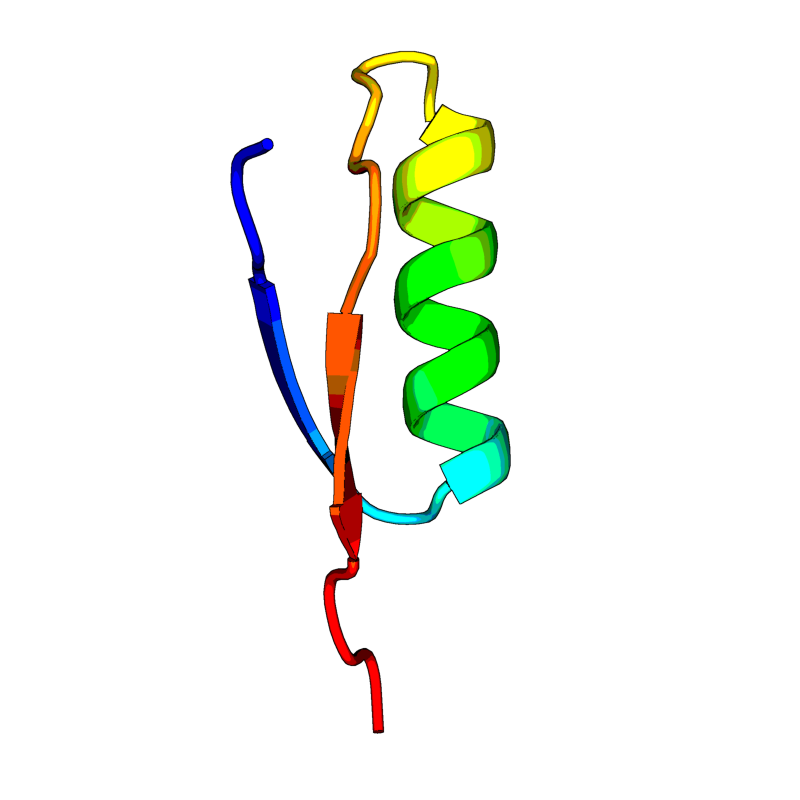 [QHI42199.pdb] Estimate TM-score=0.69 |
ORF10. Dubious/uncertain protein. It is currently unclear whether this region translates into a functional protein. The respective region in related beta-coronaviruses, such as SARS-CoV, does not correspond to translated peptide. |
NA(range:NA); |
| lineage | Structure models for close state | Structure models for open state | Protein names and sequence |
| A (Ref) |
 [close.pdb] |
 [open.pdb] |
Chain A, B, C: Spike (sequence). Chain D, E, F: ACE2. |
| B.1.1.7 (Alpha, 20I) |
 [close.pdb] |
 [open.pdb] |
Chain A, B, C: Spike (sequence). Chain D, E, F: ACE2. |
| B.1.351 (Beta, 20H) |
 [close.pdb] |
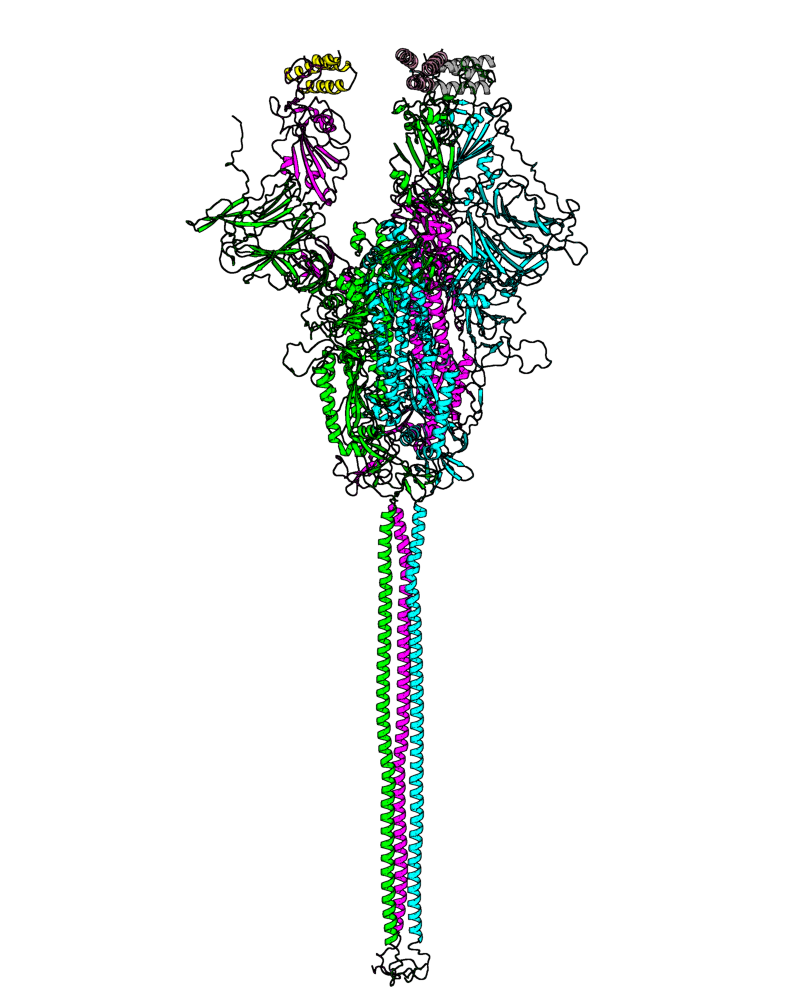 [open.pdb] |
Chain A, B, C: Spike (sequence). Chain D, E, F: ACE2. |
| P.1 (Gamma, 20J) |
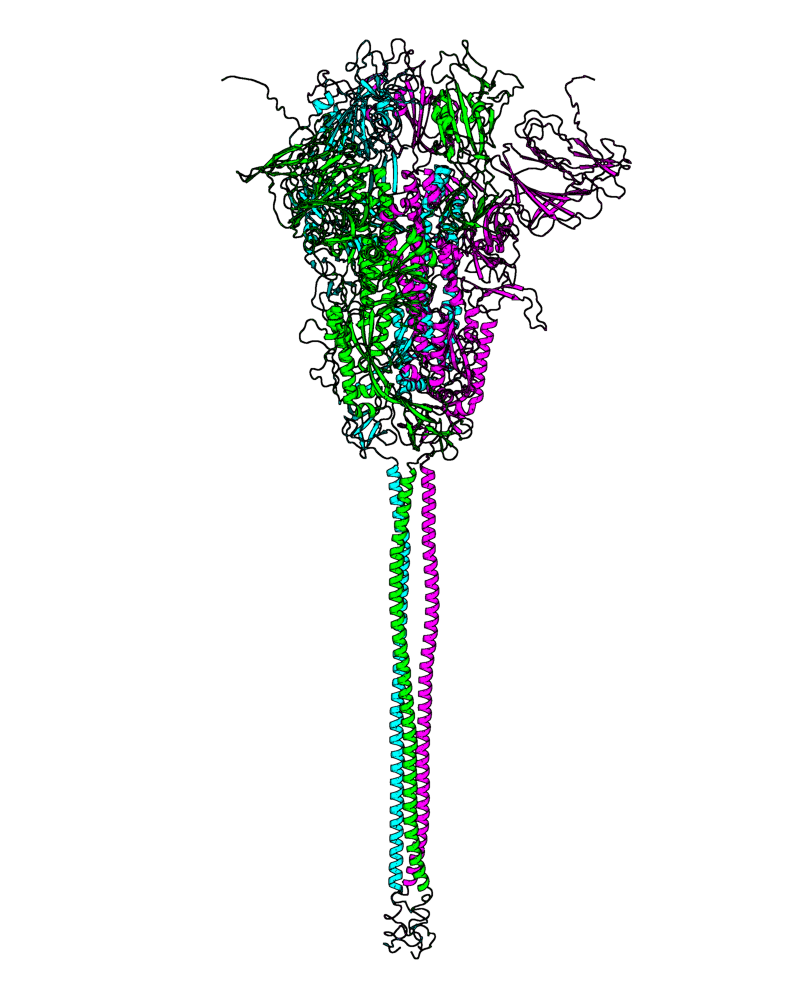 [close.pdb] |
 [open.pdb] |
Chain A, B, C: Spike (sequence). Chain D, E, F: ACE2. |
| B.1.427 (Epsilon, 21C) |
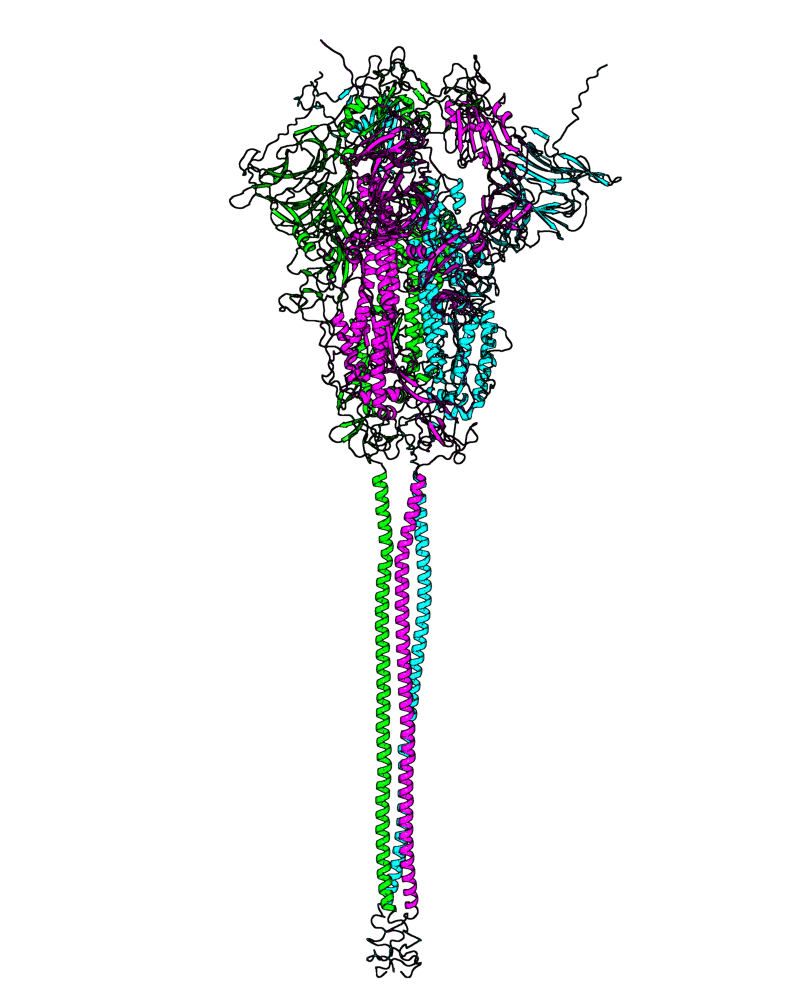 [close.pdb] |
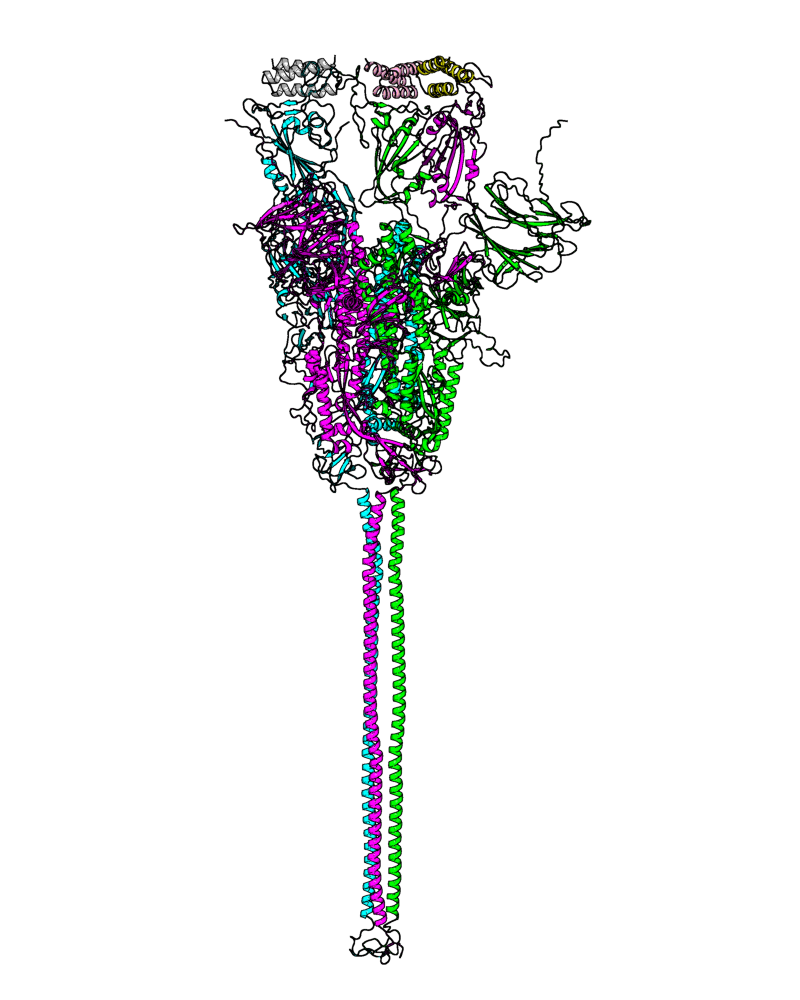 [open.pdb] |
Chain A, B, C: Spike (sequence). Chain D, E, F: ACE2. |
| B.1.429 (Epsilon, 21C) |
 [close.pdb] |
 [open.pdb] |
Chain A, B, C: Spike (sequence). Chain D, E, F: ACE2. |
| B.1.526 (Iota, 21F) |
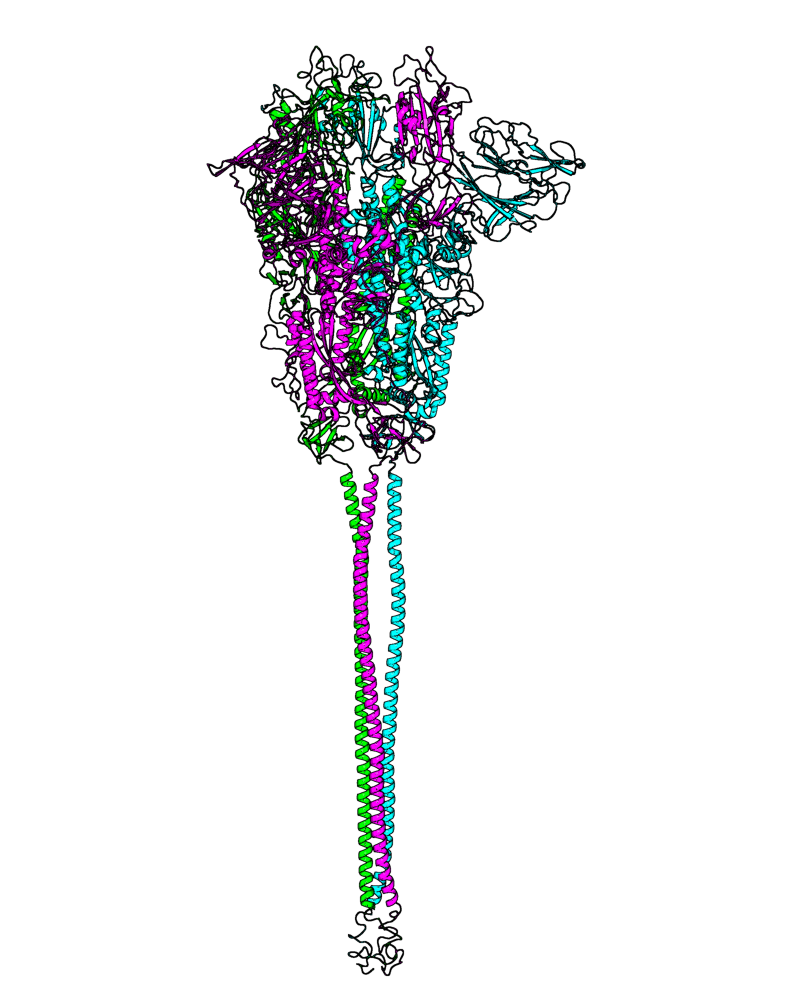 [close.pdb] |
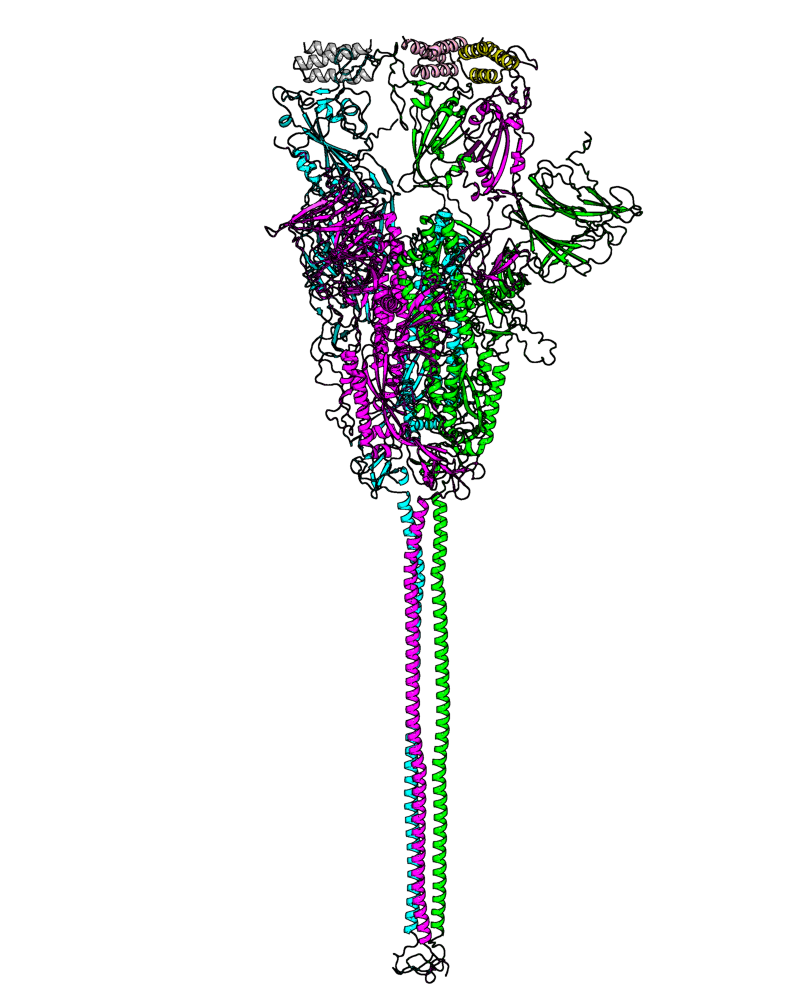 [open.pdb] |
Chain A, B, C: Spike (sequence). Chain D, E, F: ACE2. |
| B.1.177 (20E, EU1) |
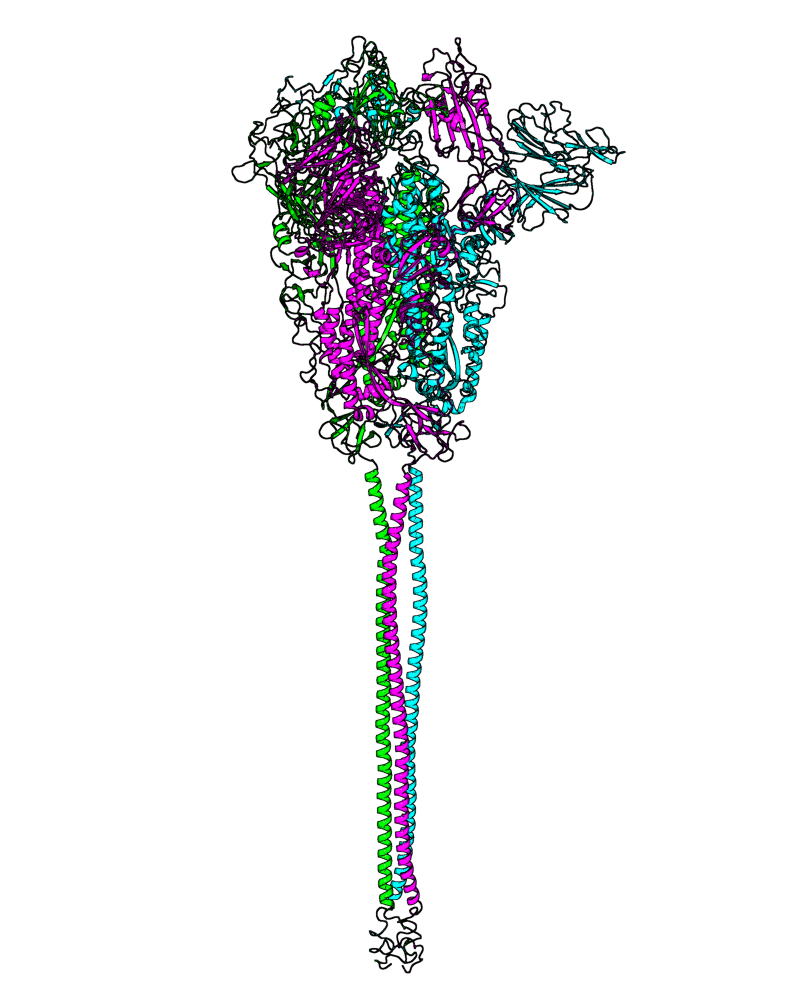 [close.pdb] |
 [open.pdb] |
Chain A, B, C: Spike (sequence). Chain D, E, F: ACE2. |
| B.1.160 (20A, EU2) |
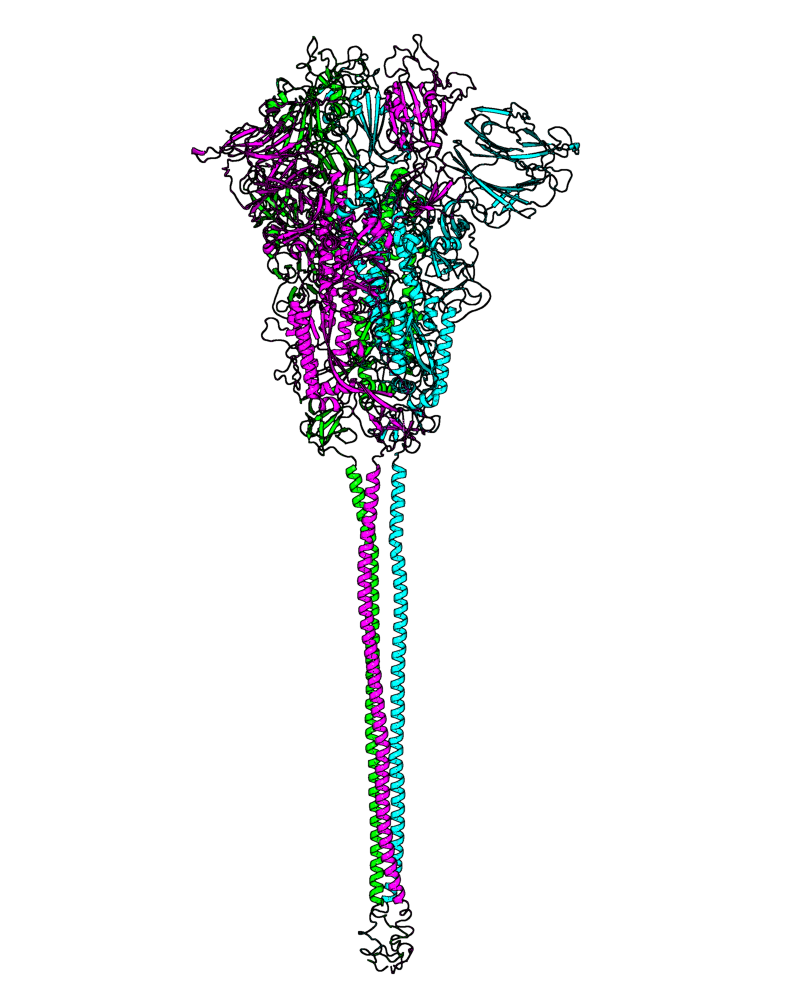 [close.pdb] |
 [open.pdb] |
Chain A, B, C: Spike (sequence). Chain D, E, F: ACE2. |
| B.1.221 (20A, S:98F) |
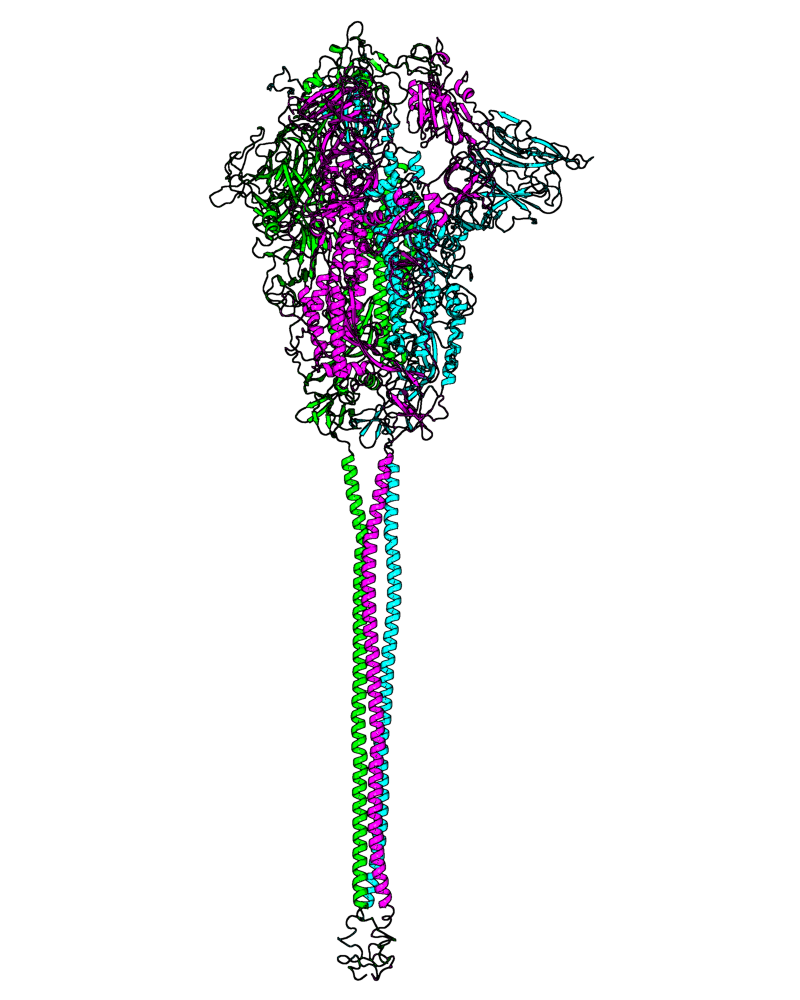 [close.pdb] |
 [open.pdb] |
Chain A, B, C: Spike (sequence). Chain D, E, F: ACE2. |
| B.1.1.277 (20B, S:626S) |
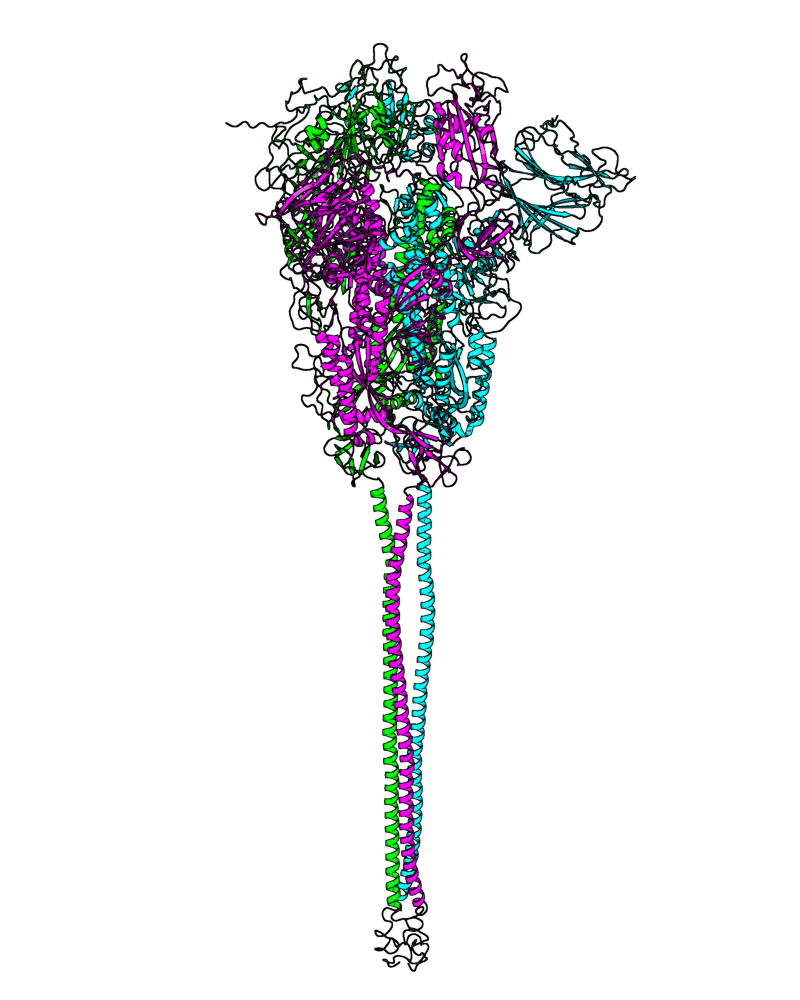 [close.pdb] |
 [open.pdb] |
Chain A, B, C: Spike (sequence). Chain D, E, F: ACE2. |
| B.1.1.302 (20B, S:1122L) |
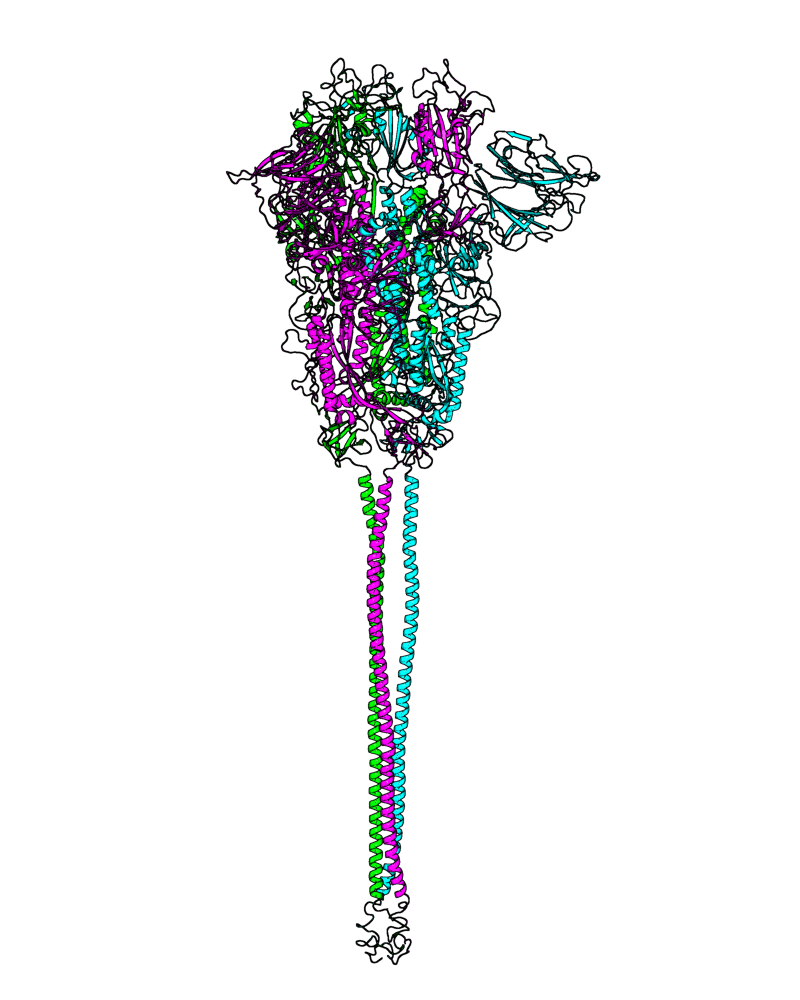 [close.pdb] |
 [open.pdb] |
Chain A, B, C: Spike (sequence). Chain D, E, F: ACE2. |



yangzhanglab![]() umich.edu
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
umich.edu
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218