

You can:
| Name | Muscarinic acetylcholine receptor M2 |
|---|---|
| Species | Rattus norvegicus (Rat) |
| Gene | Chrm2 |
| Synonym | cholinergic receptor cholinergic receptor, muscarinic 2 AChR M2 Chrm-2 M2 muscarinic acetylcholine receptor [ Show all ] |
| Disease | N/A for non-human GPCRs |
| Length | 466 |
| Amino acid sequence | MNNSTNSSNNGLAITSPYKTFEVVFIVLVAGSLSLVTIIGNILVMVSIKVNRHLQTVNNYFLFSLACADLIIGVFSMNLYTLYTVIGYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVKRTTKMAGMMIAAAWVLSFILWAPAILFWQFIVGVRTVEDGECYIQFFSNAAVTFGTAIAAFYLPVIIMTVLYWHISRASKSRIKKEKKEPVANQDPVSPSLVQGRIVKPNNNNMPGGDGGLEHNKIQNGKAPRDGVTENCVQGEEKESSNDSTSVSAVASNMRDDEITQDENTVSTSLGHSRDDNSKQTCIKIVTKAQKGDVCTPTSTTVELVGSSGQNGDEKQNIVARKIVKMTKQPAKKKPPPSREKKVTRTILAILLAFIITWAPYNVMVLINTFCAPCIPNTVWTIGYWLCYINSTINPACYALCNATFKKTFKHLLMCHYKNIGATR |
| UniProt | P10980 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | N/A |
| 3D structure model | No available structures or models |
| BioLiP | N/A |
| Therapeutic Target Database | N/A |
| ChEMBL | CHEMBL309 |
| IUPHAR | 14 |
| DrugBank | N/A |
| Name | arecoline |
|---|---|
| Molecular formula | C8H13NO2 |
| IUPAC name | methyl 1-methyl-3,6-dihydro-2H-pyridine-5-carboxylate |
| Molecular weight | 155.197 |
| Hydrogen bond acceptor | 3 |
| Hydrogen bond donor | 0 |
| XlogP | 0.3 |
| Synonyms | cid_9301 Spectrum5_001316 DTXSID3022617 ZINC52541469 KBio2_000435 [ Show all ] |
| Inchi Key | HJJPJSXJAXAIPN-UHFFFAOYSA-N |
| Inchi ID | InChI=1S/C8H13NO2/c1-9-5-3-4-7(6-9)8(10)11-2/h4H,3,5-6H2,1-2H3 |
| PubChem CID | 2230 |
| ChEMBL | CHEMBL7303 |
| IUPHAR | 296 |
| BindingDB | 46858 |
| DrugBank | DB04365 |
Structure | 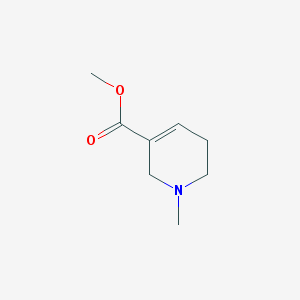 |
| Lipinski's druglikeness | This ligand satisfies Lipinski's rule of five. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| IC50 | 590.0 nM | PMID3385727 | BindingDB,ChEMBL |
| IC50 | 9150.0 nM | , Bioorg. Med. Chem. Lett., (1991) 1:3:147 | BindingDB,ChEMBL |
| IC50 | 11000.0 nM | PMID3385727 | BindingDB,ChEMBL |
| Ki | 40.0 nM | PMID9622546 | BindingDB,ChEMBL |
| Ki | 1000.0 nM | PMID4151898 | BindingDB |
| Ki | 5270.0 nM | PMID1310135 | BindingDB |
zhanglab![]() zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218