

You can:
| Name | Mu-type opioid receptor |
|---|---|
| Species | Rattus norvegicus (Rat) |
| Gene | Oprm1 |
| Synonym | Opioid receptor B opioid receptor OP3 mu receptor MOR-1 [ Show all ] |
| Disease | N/A for non-human GPCRs |
| Length | 398 |
| Amino acid sequence | MDSSTGPGNTSDCSDPLAQASCSPAPGSWLNLSHVDGNQSDPCGLNRTGLGGNDSLCPQTGSPSMVTAITIMALYSIVCVVGLFGNFLVMYVIVRYTKMKTATNIYIFNLALADALATSTLPFQSVNYLMGTWPFGTILCKIVISIDYYNMFTSIFTLCTMSVDRYIAVCHPVKALDFRTPRNAKIVNVCNWILSSAIGLPVMFMATTKYRQGSIDCTLTFSHPTWYWENLLKICVFIFAFIMPVLIITVCYGLMILRLKSVRMLSGSKEKDRNLRRITRMVLVVVAVFIVCWTPIHIYVIIKALITIPETTFQTVSWHFCIALGYTNSCLNPVLYAFLDENFKRCFREFCIPTSSTIEQQNSTRVRQNTREHPSTANTVDRTNHQLENLEAETAPLP |
| UniProt | P33535 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | N/A |
| 3D structure model | No available structures or models |
| BioLiP | N/A |
| Therapeutic Target Database | N/A |
| ChEMBL | CHEMBL270 |
| IUPHAR | 319 |
| DrugBank | N/A |
| Name | DAMGO |
|---|---|
| Molecular formula | C26H35N5O6 |
| IUPAC name | (2S)-2-amino-N-[(2R)-1-[[2-[[(2S)-1-(2-hydroxyethylamino)-1-oxo-3-phenylpropan-2-yl]-methylamino]-2-oxoethyl]amino]-1-oxopropan-2-yl]-3-(4-hydroxyphenyl)propanamide |
| Molecular weight | 513.595 |
| Hydrogen bond acceptor | 7 |
| Hydrogen bond donor | 6 |
| XlogP | 0.2 |
| Synonyms | ZINC53092426 (2S)-2-amino-N-[(1R)-2-[[2-[[(1S)-1-benzyl-2-(2-hydroxyethylamino)-2-oxo-ethyl]-methyl-amino]-2-oxo-ethyl]amino]-1-methyl-2-oxo-ethyl]-3-(4-hydroxyphenyl)propanamide 2-Ala-4-mephe-5-gly-enkephalin AC1Q29AA CHEBI:272 [ Show all ] |
| Inchi Key | HPZJMUBDEAMBFI-WTNAPCKOSA-N |
| Inchi ID | InChI=1S/C26H35N5O6/c1-17(30-25(36)21(27)14-19-8-10-20(33)11-9-19)24(35)29-16-23(34)31(2)22(26(37)28-12-13-32)15-18-6-4-3-5-7-18/h3-11,17,21-22,32-33H,12-16,27H2,1-2H3,(H,28,37)(H,29,35)(H,30,36)/t17-,21+,22+/m1/s1 |
| PubChem CID | 5462471 |
| ChEMBL | CHEMBL38874 |
| IUPHAR | 1647 |
| BindingDB | 21015 |
| DrugBank | N/A |
Structure | 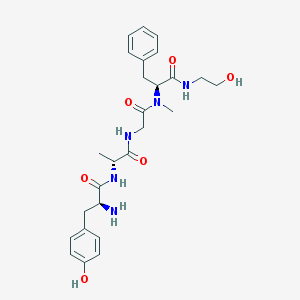 |
| Lipinski's druglikeness | This ligand has more than 5 hydrogen bond donor. This ligand is heavier than 500 daltons. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| Activity | 76.0 % | PMID19640720 | ChEMBL |
| Bmax | 1207.0 fM mg-1 | PMID10866377 | ChEMBL |
| EC50 | 36.0 nM | PMID20560643, PMID16509592 | BindingDB |
| EC50 | 36.31 nM | PMID19762245, PMID16686530, PMID17516639, PMID20617791, PMID20560643, PMID21366266, PMID21128594, PMID16509592 | ChEMBL |
| EC50 | 37.0 nM | PMID18821747, PMID17927164, PMID19762245, PMID16686530, PMID17516639, PMID20617791, PMID20560643, PMID21366266, PMID21128594, PMID16509592, PMID18266313 | BindingDB,ChEMBL |
| EC50 | 40.0 nM | PMID18821747 | BindingDB |
| EC50 | 81.0 nM | PMID25221662 | BindingDB,ChEMBL |
| EC50 | 145.0 nM | PMID18572932 | BindingDB,ChEMBL |
| EC50 | 214.0 nM | PMID21287991 | BindingDB,ChEMBL |
| EC50 | 485.0 nM | PMID22957923 | BindingDB,ChEMBL |
| Emax | 119.0 % | PMID21287991 | ChEMBL |
| Emax | 150.0 % | PMID18821747, PMID19762245, PMID16686530, PMID17516639, PMID20617791, PMID20560643, PMID21366266, PMID21128594, PMID16509592, PMID18266313 | ChEMBL |
| Emax | 169.0 % | PMID18062664 | ChEMBL |
| Emax | 170.0 % | PMID18572932 | ChEMBL |
| Emax | 173.0 % | PMID22957923 | ChEMBL |
| Emax | 178.0 % | PMID21287991 | ChEMBL |
| Emax | 465.2 % | PMID25221662 | ChEMBL |
| IC50 | 1.01 nM | PMID19640720 | BindingDB,ChEMBL |
| IC50 | 1.13 nM | PMID17395470 | BindingDB,ChEMBL |
| IC50 | 3.0 nM | PMID9873480 | BindingDB,ChEMBL |
| IC50 | 3.5 nM | PMID10543877 | BindingDB,ChEMBL |
| IC50 | 9.89 nM | PMID11133084 | ChEMBL |
| IC50 | 9.9 nM | PMID11133084, PMID12036366, PMID15456262 | BindingDB,ChEMBL |
| IC50 | 9.9 nM | PMID15456262 | BindingDB |
| IC50 | 36.31 nM | PMID19762245 | ChEMBL |
| Kd | 0.5 nM | PMID18266313 | BindingDB |
| Kd | 0.85 nM | PMID16686530, PMID17927164 | BindingDB |
| Ki | 0.0024 nM | PMID18062664 | BindingDB,ChEMBL |
| Ki | 0.365 nM | PMID7562497 | BindingDB |
| Ki | 0.53 nM | PMID10866377 | ChEMBL |
| Ki | 0.53 nM | PMID10866377 | BindingDB |
| Ki | 0.62 nM | PMID9694962 | BindingDB |
| Ki | 0.66 nM | PMID1849997 | ChEMBL |
| Ki | 0.66 nM | PMID1849997 | BindingDB |
| Ki | 0.85 nM | PMID20617791 | ChEMBL |
| Ki | 0.85 nM | PMID20617791 | BindingDB |
| Ki | 0.87 nM | PMID20599386 | ChEMBL |
| Ki | 0.87 nM | PMID20599386 | BindingDB |
| Ki | 1.0 nM | PMID25221662 | ChEMBL |
| Ki | 1.0 nM | PMID25221662 | BindingDB |
| Ki | 1.1 nM | PMID10585536 | BindingDB |
| Ki | 1.16 nM | PMID9700760 | BindingDB |
| Ki | 1.2 nM | PMID2537427 | BindingDB |
| Ki | 1.22 nM | PMID2537427 | ChEMBL |
| Ki | 1.29 nM | PMID9700760 | BindingDB |
| Ki | 1.49 nM | PMID9694962 | BindingDB |
| Ki | 1.6 nM | PMID11133084, PMID12036366, PMID15456262 | BindingDB,ChEMBL |
| Ki | 1.64 nM | PMID11133084 | ChEMBL |
| Ki | 1.7 nM | PMID23974016 | ChEMBL |
| Ki | 1.9 nM | PMID3033464 | BindingDB |
| Ki | 1.99526 nM | PMID8114680 | IUPHAR |
| Ki | 3.9 nM | PMID2828622, PMID1714957 | BindingDB,ChEMBL |
| Ki | 4.0 nM | PMID8064796 | BindingDB,ChEMBL |
| Ki | 6.1 nM | PMID14998329 | BindingDB,ChEMBL |
| Ki | 16.0 nM | PMID19762245 | BindingDB,ChEMBL |
| Log10ED50 | 6.71 - | PMID18062664 | ChEMBL |
| logEC50 | -7.4 - | PMID18266313 | ChEMBL |
| logIC50 | 8.62 - | PMID23974016 | ChEMBL |
| pEC50 | 7.4 nM | PMID18821747 | ChEMBL |
| Relative potency | 7.73 - | PMID2537427 | ChEMBL |
zhanglab![]() zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218