

You can:
| Name | Mu-type opioid receptor |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | OPRM1 |
| Synonym | hMOP M-OR-1 MOP opioid receptor, mu 1 opioid receptor [ Show all ] |
| Disease | Diarrhea Inflammatory disease Pain Major depressive disorder Migraine [ Show all ] |
| Length | 400 |
| Amino acid sequence | MDSSAAPTNASNCTDALAYSSCSPAPSPGSWVNLSHLDGNLSDPCGPNRTDLGGRDSLCPPTGSPSMITAITIMALYSIVCVVGLFGNFLVMYVIVRYTKMKTATNIYIFNLALADALATSTLPFQSVNYLMGTWPFGTILCKIVISIDYYNMFTSIFTLCTMSVDRYIAVCHPVKALDFRTPRNAKIINVCNWILSSAIGLPVMFMATTKYRQGSIDCTLTFSHPTWYWENLLKICVFIFAFIMPVLIITVCYGLMILRLKSVRMLSGSKEKDRNLRRITRMVLVVVAVFIVCWTPIHIYVIIKALVTIPETTFQTVSWHFCIALGYTNSCLNPVLYAFLDENFKRCFREFCIPTSSNIEQQNSTRIRQNTRDHPSTANTVDRTNHQLENLEAETAPLP |
| UniProt | P35372 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | P35372 |
| 3D structure model | This predicted structure model is from GPCR-EXP P35372. |
| BioLiP | N/A |
| Therapeutic Target Database | T47768 |
| ChEMBL | CHEMBL233 |
| IUPHAR | 319 |
| DrugBank | BE0000770 |
| Name | DAMGO |
|---|---|
| Molecular formula | C26H35N5O6 |
| IUPAC name | (2S)-2-amino-N-[(2R)-1-[[2-[[(2S)-1-(2-hydroxyethylamino)-1-oxo-3-phenylpropan-2-yl]-methylamino]-2-oxoethyl]amino]-1-oxopropan-2-yl]-3-(4-hydroxyphenyl)propanamide |
| Molecular weight | 513.595 |
| Hydrogen bond acceptor | 7 |
| Hydrogen bond donor | 6 |
| XlogP | 0.2 |
| Synonyms | CHEMBL38874 H-Tyr-D-Ala-Gly-MePhe-Gly-ol RX 783006 Tyr-D-Ala-Gly-N-Me-Phe-Gly-ol [D-Ala2, NMe-Phe4, Gly-ol5]-enkephalin [ Show all ] |
| Inchi Key | HPZJMUBDEAMBFI-WTNAPCKOSA-N |
| Inchi ID | InChI=1S/C26H35N5O6/c1-17(30-25(36)21(27)14-19-8-10-20(33)11-9-19)24(35)29-16-23(34)31(2)22(26(37)28-12-13-32)15-18-6-4-3-5-7-18/h3-11,17,21-22,32-33H,12-16,27H2,1-2H3,(H,28,37)(H,29,35)(H,30,36)/t17-,21+,22+/m1/s1 |
| PubChem CID | 5462471 |
| ChEMBL | CHEMBL38874 |
| IUPHAR | 1647 |
| BindingDB | 21015 |
| DrugBank | N/A |
Structure | 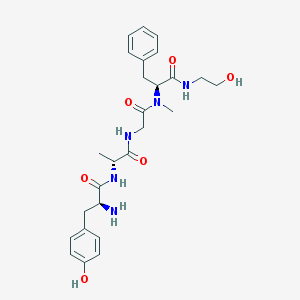 |
| Lipinski's druglikeness | This ligand has more than 5 hydrogen bond donor. This ligand is heavier than 500 daltons. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| Activity | 100.0 % | PMID21866885 | ChEMBL |
| Activity | 366.0 % | PMID19217280 | ChEMBL |
| Bmax | 9.2 pmol | PMID19113864 | ChEMBL |
| EC50 | 1.0 nM | PMID17616524 | ChEMBL |
| EC50 | 1.7 nM | PMID18637671 | BindingDB,ChEMBL |
| EC50 | 3.0 nM | PMID22724433 | BindingDB |
| EC50 | 3.0 nM | PMID23477419 | BindingDB |
| EC50 | 3.04 nM | PMID23477419, PMID22724433 | ChEMBL |
| EC50 | 3.18 nM | PMID21978284 | ChEMBL |
| EC50 | 3.2 nM | PMID21978284 | BindingDB |
| EC50 | 7.5 nM | PMID25783191 | BindingDB,ChEMBL |
| EC50 | 7.57 nM | PMID25248680 | ChEMBL |
| EC50 | 7.6 nM | PMID25248680 | BindingDB |
| EC50 | 8.2 nM | PMID19683449 | BindingDB,ChEMBL |
| EC50 | 9.6 nM | PMID25783191 | BindingDB |
| EC50 | 9.61 nM | PMID25783191 | ChEMBL |
| EC50 | 13.7 nM | PMID12672258 | ChEMBL |
| EC50 | 14.0 nM | PMID12672258 | BindingDB |
| EC50 | 15.0 nM | PMID23880538 | ChEMBL |
| EC50 | 30.0 nM | PMID27096047 | BindingDB,ChEMBL |
| EC50 | 32.6 nM | PMID24657054 | ChEMBL |
| EC50 | 33.0 nM | PMID24657054 | BindingDB |
| EC50 | 35.0 nM | PMID21866885 | BindingDB |
| EC50 | 35.3 nM | PMID21866885 | ChEMBL |
| EC50 | 40.0 nM | PMID18380425 | BindingDB,ChEMBL |
| EC50 | 45.0 nM | PMID19217280, PMID19199782 | BindingDB |
| EC50 | 45.06 nM | PMID19199782 | ChEMBL |
| EC50 | 45.1 nM | PMID19217280 | ChEMBL |
| EC50 | 55.0 nM | PMID17935988, PMID19282177, PMID23434225, PMID16942039, PMID19027293, PMID19091564, PMID22439881, PMID18417347, PMID23142613 | BindingDB,ChEMBL |
| EC50 | 110.0 nM | PMID17276685, PMID17407276, PMID16392810, PMID17433695, PMID21351746 | BindingDB,ChEMBL |
| EC50 | 202.0 nM | PMID15380196 | BindingDB,ChEMBL |
| EC50 | 378.0 nM | PMID24613457 | BindingDB,ChEMBL |
| ED50 | 34.0 nM | PMID23618710 | ChEMBL |
| ED50 | 42.0 nM | PMID19627147, PMID17625813, PMID20055417 | ChEMBL |
| Emax | 73.0 % | PMID21978284 | ChEMBL |
| Emax | 95.0 % | PMID15380196 | ChEMBL |
| Emax | 98.1 % | PMID23477419 | ChEMBL |
| Emax | 98.14 % | PMID22724433 | ChEMBL |
| Emax | 100.0 % | PMID23618710, PMID17625813, PMID20055417 | ChEMBL |
| Emax | 116.0 % | PMID17935988, PMID19027293, PMID16942039, PMID23434225, PMID19091564, PMID18417347, PMID23142613 | ChEMBL |
| Emax | 120.0 % | PMID19282177, PMID17433695, PMID17407276, PMID21351746, PMID16392810, PMID17276685 | ChEMBL |
| Emax | 124.0 % | PMID19683449 | ChEMBL |
| Emax | 130.0 % | PMID22439881 | ChEMBL |
| Emax | 225.0 % | PMID27096047 | ChEMBL |
| Emax | 366.5 % | PMID19199782 | ChEMBL |
| Emax | 389.0 % | PMID18062664 | ChEMBL |
| IC50 | 0.43 nM | PMID26988801, PMID27876250 | ChEMBL |
| IC50 | 0.68 nM | PMID23466604 | ChEMBL |
| IC50 | 0.74 nM | PMID23582449 | ChEMBL |
| IC50 | 1.1 nM | PMID18983139 | ChEMBL |
| IC50 | 1.6 nM | PMID11906279 | BindingDB,ChEMBL |
| IC50 | 2.0 nM | PMID20426456 | BindingDB,ChEMBL |
| IC50 | 2.5 nM | PMID26789491, PMID18588282 | BindingDB,ChEMBL |
| IC50 | 3.18 nM | PMID20599386 | ChEMBL |
| IC50 | 3.2 nM | PMID20599386 | BindingDB |
| IC50 | 4.3 nM | PMID26988801, PMID27876250 | BindingDB |
| IC50 | 5.0 nM | PMID25087049 | ChEMBL |
| IC50 | 5.0 nM | PMID25087049 | BindingDB |
| IC50 | 20.0 nM | PMID23403082 | ChEMBL |
| Imax | 73.0 % | PMID20599386 | ChEMBL |
| Inhibition | 4.0 % | PMID23403082 | ChEMBL |
| Inhibition | 96.0 % | PMID23880538 | ChEMBL |
| Ki | 0.28 nM | PMID23466604 | ChEMBL |
| Ki | 0.44 nM | PMID18983139 | ChEMBL |
| Ki | 0.45 nM | PMID21621410 | ChEMBL |
| Ki | 0.45 nM | PMID21621410 | BindingDB |
| Ki | 0.5 nM | PMID12672258 | BindingDB |
| Ki | 0.5 nM | PMID12672258, PMID9686407 | BindingDB,ChEMBL |
| Ki | 0.501187 nM | PMID9686407, PMID6263640 | IUPHAR |
| Ki | 0.51 nM | PMID7932177 | BindingDB |
| Ki | 0.53 nM | PMID12699761 | ChEMBL |
| Ki | 0.53 nM | PMID12699761 | BindingDB |
| Ki | 0.56 nM | PMID26125201 | ChEMBL |
| Ki | 0.56 nM | PMID26125201 | BindingDB |
| Ki | 0.9 nM | PMID19683449, PMID23587424 | ChEMBL |
| Ki | 0.9 nM | PMID19683449 | BindingDB |
| Ki | 1.2 nM | PMID25599950, PMID23618710, PMID22341895, PMID23880358 | BindingDB,ChEMBL |
| Ki | 1.259 nM | PMID26632862 | ChEMBL |
| Ki | 1.3 nM | PMID26632862 | BindingDB |
| Ki | 1.4 nM | PMID7815359 | BindingDB |
| Ki | 1.5 nM | PMID22995061, PMID25051243 | BindingDB,ChEMBL |
| Ki | 1.55 nM | PMID21621410 | ChEMBL |
| Ki | 1.59 nM | PMID21866885 | ChEMBL |
| Ki | 1.6 nM | PMID21866885, PMID21621410 | BindingDB |
| Ki | 2.0 nM | PMID8114680 | BindingDB |
| Ki | 2.3 nM | PMID25193297 | BindingDB,ChEMBL |
| Ki | 2.65 nM | PMID12513698 | BindingDB |
| Ki | 2.96 nM | PMID24657054 | ChEMBL |
| Ki | 3.0 nM | PMID24657054 | BindingDB |
| Ki | 3.3 nM | PMID21621410 | BindingDB |
| Ki | 3.34 nM | PMID21621410 | ChEMBL |
| Ki | 3.715 nM | PMID17490886 | ChEMBL |
| Ki | 3.8 nM | PMID19836950 | BindingDB,ChEMBL |
| Ki | 6.7 nM | PMID19113864 | BindingDB,ChEMBL |
| Ki | 8.1 nM | PMID23403082 | ChEMBL |
| Ki | 14.0 nM | PMID9651168 | BindingDB,ChEMBL |
| Ki | 36.7 nM | PMID15380196 | ChEMBL |
| Ki | 37.0 nM | PMID15380196 | BindingDB |
| Ki | 87.0 nM | PMID26632862 | BindingDB |
| Ki | 87.1 nM | PMID26632862 | ChEMBL |
| Ki | 2187.76 nM | PMID26632862 | ChEMBL |
| Ki | 2188.0 nM | PMID26632862 | BindingDB |
| Log10ED50 | 7.18 - | PMID18062664 | ChEMBL |
| Ratio | 0.8 - | PMID24613457 | ChEMBL |
| Stimulation | 100.0 % | PMID12672258 | ChEMBL |
zhanglab![]() zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218