

You can:
| Name | 5-hydroxytryptamine receptor 7 |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | HTR7 |
| Synonym | 5-HT-7 Serotonin receptor 7 5-HT-X 5-HT1Y 5-HT7 [ Show all ] |
| Disease | Sleep disorders Schizophrenia Major depressive disorder |
| Length | 479 |
| Amino acid sequence | MMDVNSSGRPDLYGHLRSFLLPEVGRGLPDLSPDGGADPVAGSWAPHLLSEVTASPAPTWDAPPDNASGCGEQINYGRVEKVVIGSILTLITLLTIAGNCLVVISVCFVKKLRQPSNYLIVSLALADLSVAVAVMPFVSVTDLIGGKWIFGHFFCNVFIAMDVMCCTASIMTLCVISIDRYLGITRPLTYPVRQNGKCMAKMILSVWLLSASITLPPLFGWAQNVNDDKVCLISQDFGYTIYSTAVAFYIPMSVMLFMYYQIYKAARKSAAKHKFPGFPRVEPDSVIALNGIVKLQKEVEECANLSRLLKHERKNISIFKREQKAATTLGIIVGAFTVCWLPFFLLSTARPFICGTSCSCIPLWVERTFLWLGYANSLINPFIYAFFNRDLRTTYRSLLQCQYRNINRKLSAAGMHEALKLAERPERPEFVLRACTRRVLLRPEKRPPVSVWVLQSPDHHNWLADKMLTTVEKKVMIHD |
| UniProt | P34969 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | P34969 |
| 3D structure model | This predicted structure model is from GPCR-EXP P34969. |
| BioLiP | N/A |
| Therapeutic Target Database | T79062 |
| ChEMBL | CHEMBL3155 |
| IUPHAR | 12 |
| DrugBank | BE0000650, BE0004862 |
| Name | SB-269970 |
|---|---|
| Molecular formula | C18H28N2O3S |
| IUPAC name | 3-[(2R)-2-[2-(4-methylpiperidin-1-yl)ethyl]pyrrolidin-1-yl]sulfonylphenol |
| Molecular weight | 352.493 |
| Hydrogen bond acceptor | 5 |
| Hydrogen bond donor | 1 |
| XlogP | 2.9 |
| Synonyms | (R)-3-((2-(2-(4-Methylpiperidin-1-yl)ethyl)pyrrolidin-1-yl)sulfonyl)phenol 3-{(R)-2-[2-(4-Methyl-piperidin-1-yl)-ethyl]-pyrrolidine-1-sulfonyl}-phenol AC1O7H0E CCG-205136 FT-0771258 [ Show all ] |
| Inchi Key | HWKROQUZSKPIKQ-MRXNPFEDSA-N |
| Inchi ID | InChI=1S/C18H28N2O3S/c1-15-7-11-19(12-8-15)13-9-16-4-3-10-20(16)24(22,23)18-6-2-5-17(21)14-18/h2,5-6,14-16,21H,3-4,7-13H2,1H3/t16-/m1/s1 |
| PubChem CID | 6604889 |
| ChEMBL | CHEMBL282199 |
| IUPHAR | 3233, 3247 |
| BindingDB | 50098551 |
| DrugBank | DB13988 |
Structure | 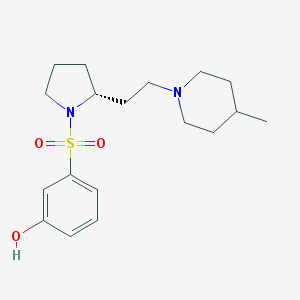 |
| Lipinski's druglikeness | This ligand satisfies Lipinski's rule of five. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| N/A | N/A | DrugBank | |
| Activity | <95.0 % | PMID18361484 | ChEMBL |
| Activity | 80.0 % | PMID18361484 | ChEMBL |
| IC50 | 3.7 nM | PMID27475109 | BindingDB |
| IC50 | 3.71 nM | PMID27475109 | ChEMBL |
| IC50 | 3.715 nM | PMID27475109 | ChEMBL |
| Imax | 100.0 % | PMID27475109 | ChEMBL |
| Inhibition | 97.0 % | PMID28063784 | ChEMBL |
| Inhibition | 102.0 % | PMID25127461, PMID26852005 | ChEMBL |
| Inhibition | 104.0 % | PMID23810425 | ChEMBL |
| Kb | 1.2 nM | PMID24805037 | ChEMBL |
| Kd | 1.2 nM | PMID10807680 | IUPHAR |
| Kd | 9.7 nM | PMID21620533 | BindingDB,ChEMBL |
| Ki | 0.3388 nM | PMID26852005 | ChEMBL |
| Ki | 0.338844 nM | PMID26852005 | BindingDB |
| Ki | 0.34 nM | PMID25127461, PMID27475109, PMID26852005 | ChEMBL |
| Ki | 0.34 nM | PMID25127461, PMID27475109, PMID26852005 | BindingDB |
| Ki | 0.71 nM | PMID28063784, PMID25759032 | BindingDB |
| Ki | 0.71 nM | PMID28063784, PMID25759032 | ChEMBL |
| Ki | 1.0 nM | PMID21159507, PMID12161155 | BindingDB,ChEMBL |
| Ki | 1.2 nM | PMID22001327, PMID22926225, PMID19560916 | BindingDB,ChEMBL |
| Ki | 1.25893 - 2.51189 nM | PMID10807680 | IUPHAR |
| Ki | 1.259 nM | PMID10669560, PMID14667218, PMID27717652 | ChEMBL |
| Ki | 1.26 nM | PMID25128671, PMID23541835, PMID10669560 | BindingDB,ChEMBL |
| Ki | 1.3 nM | PMID25128671, PMID23541835, PMID12392747, PMID12825922, PMID27717652 | BindingDB,ChEMBL |
| pKb | 8.3 - | PMID12825922 | ChEMBL |
zhanglab![]() zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218