

You can:
| Name | Beta-2 adrenergic receptor |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | ADRB2 |
| Synonym | beta-2 adrenergic receptor Gpcr7 beta2-adrenoceptor Adrb-2 ADRB2R [ Show all ] |
| Disease | Premature labour Premature ejaculation Obesity Neurogenic bladder dysfunction Hypertension [ Show all ] |
| Length | 413 |
| Amino acid sequence | MGQPGNGSAFLLAPNGSHAPDHDVTQERDEVWVVGMGIVMSLIVLAIVFGNVLVITAIAKFERLQTVTNYFITSLACADLVMGLAVVPFGAAHILMKMWTFGNFWCEFWTSIDVLCVTASIETLCVIAVDRYFAITSPFKYQSLLTKNKARVIILMVWIVSGLTSFLPIQMHWYRATHQEAINCYANETCCDFFTNQAYAIASSIVSFYVPLVIMVFVYSRVFQEAKRQLQKIDKSEGRFHVQNLSQVEQDGRTGHGLRRSSKFCLKEHKALKTLGIIMGTFTLCWLPFFIVNIVHVIQDNLIRKEVYILLNWIGYVNSGFNPLIYCRSPDFRIAFQELLCLRRSSLKAYGNGYSSNGNTGEQSGYHVEQEKENKLLCEDLPGTEDFVGHQGTVPSDNIDSQGRNCSTNDSLL |
| UniProt | P07550 |
| Protein Data Bank | 3nya, 3ny9, 3ny8, 3d4s, 2rh1, 3pds, 4gbr, 4lde, 6mxt, 6csy, 5x7d, 5jqh, 5d6l, 5d5b, 5d5a, 4ldo, 4ldl, 4qkx |
| GPCR-HGmod model | P07550 |
| 3D structure model | This structure is from PDB ID 3nya. |
| BioLiP | BL0257082, BL0113951, BL0113950, BL0257084, BL0257085, BL0283869, BL0333729, BL0333730, BL0333731,BL0333732,BL0333733, BL0333734, BL0333735, BL0333736,BL0333737,BL0333738, BL0113952,BL0113953,BL0113954, BL0147310, BL0257081, BL0257080, BL0232997, BL0192129, BL0257083, BL0185746,BL0185747, BL0185745, BL0185743,BL0185744, BL0185742, BL0185740,BL0185741, BL0147311,BL0147312, BL0351701,BL0351703, BL0351702,BL0351704, BL0192128, BL0433200, BL0433199, BL0430930, BL0430929, BL0388810, BL0388809, BL0354449, BL0185748, BL0354450, BL0354451,BL0354452,BL0354453, BL0388807,BL0388808 |
| Therapeutic Target Database | T24555, T52522 |
| ChEMBL | CHEMBL210 |
| IUPHAR | 29 |
| DrugBank | BE0000694 |
| Name | CHEMBL67998 |
|---|---|
| Molecular formula | C33H38N4O5S |
| IUPAC name | 1-[4-[[6-[(6,7-dihydroxy-1,2,3,4-tetrahydroisoquinolin-1-yl)methyl]naphthalen-2-yl]sulfamoyl]phenyl]-3-hexylurea |
| Molecular weight | 602.75 |
| Hydrogen bond acceptor | 7 |
| Hydrogen bond donor | 6 |
| XlogP | 5.7 |
| Synonyms | BDBM50093041 N-[6-(6,7-Dihydroxy-1,2,3,4-tetrahydro-isoquinolin-1-ylmethyl)-naphthalen-2-yl]-4-(3-hexyl-ureido)-benzenesulfonamide N-[6-(6,7-Dihydroxy-1,2,3,4-tetrahydroisoquinoline-1-ylmethyl)-2-naphthyl]-4-(3-hexylureido)benzenesulfonamide |
| Inchi Key | ASCKRJRRWMTXJM-UHFFFAOYSA-N |
| Inchi ID | InChI=1S/C33H38N4O5S/c1-2-3-4-5-15-35-33(40)36-26-10-12-28(13-11-26)43(41,42)37-27-9-8-23-17-22(6-7-24(23)19-27)18-30-29-21-32(39)31(38)20-25(29)14-16-34-30/h6-13,17,19-21,30,34,37-39H,2-5,14-16,18H2,1H3,(H2,35,36,40) |
| PubChem CID | 44312949 |
| ChEMBL | CHEMBL67998 |
| IUPHAR | N/A |
| BindingDB | 50093041 |
| DrugBank | N/A |
Structure | 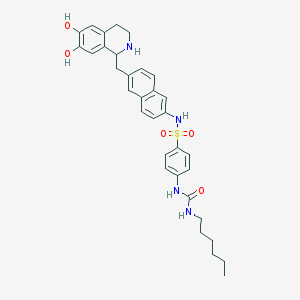 |
| Lipinski's druglikeness | This ligand has more than 5 hydrogen bond donor. This ligand is heavier than 500 daltons. This ligand has a partition coefficient log P greater than 5. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| IC50 | 300.0 nM | PMID11055339 | BindingDB,ChEMBL |
zhanglab![]() zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218