

You can:
| Name | 5-hydroxytryptamine receptor 1A |
|---|---|
| Species | Rattus norvegicus (Rat) |
| Gene | Htr1a |
| Synonym | 5-HT1A receptor 5-hydroxytryptamine (serotonin) receptor 1A, G protein-coupled 5-HT1A ADRB2RL1 ADRBRL1 [ Show all ] |
| Disease | N/A for non-human GPCRs |
| Length | 422 |
| Amino acid sequence | MDVFSFGQGNNTTASQEPFGTGGNVTSISDVTFSYQVITSLLLGTLIFCAVLGNACVVAAIALERSLQNVANYLIGSLAVTDLMVSVLVLPMAALYQVLNKWTLGQVTCDLFIALDVLCCTSSILHLCAIALDRYWAITDPIDYVNKRTPRRAAALISLTWLIGFLISIPPMLGWRTPEDRSDPDACTISKDHGYTIYSTFGAFYIPLLLMLVLYGRIFRAARFRIRKTVRKVEKKGAGTSLGTSSAPPPKKSLNGQPGSGDWRRCAENRAVGTPCTNGAVRQGDDEATLEVIEVHRVGNSKEHLPLPSESGSNSYAPACLERKNERNAEAKRKMALARERKTVKTLGIIMGTFILCWLPFFIVALVLPFCESSCHMPALLGAIINWLGYSNSLLNPVIYAYFNKDFQNAFKKIIKCKFCRR |
| UniProt | P19327 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | N/A |
| 3D structure model | No available structures or models |
| BioLiP | N/A |
| Therapeutic Target Database | N/A |
| ChEMBL | CHEMBL273 |
| IUPHAR | 1 |
| DrugBank | N/A |
| Name | 8-OH-Dpat |
|---|---|
| Molecular formula | C16H25NO |
| IUPAC name | 7-(dipropylamino)-5,6,7,8-tetrahydronaphthalen-1-ol |
| Molecular weight | 247.382 |
| Hydrogen bond acceptor | 2 |
| Hydrogen bond donor | 1 |
| XlogP | 4.1 |
| Synonyms | 1-Naphthalenol, 7-(dipropylamino)-5,6,7,8-tetrahydro- 8-OH-DPAT,(+) ASXGJMSKWNBENU-UHFFFAOYSA-N CHEBI:73364 GTPL31 [ Show all ] |
| Inchi Key | ASXGJMSKWNBENU-UHFFFAOYSA-N |
| Inchi ID | InChI=1S/C16H25NO/c1-3-10-17(11-4-2)14-9-8-13-6-5-7-16(18)15(13)12-14/h5-7,14,18H,3-4,8-12H2,1-2H3 |
| PubChem CID | 1220 |
| ChEMBL | CHEMBL56 |
| IUPHAR | 7, 31 |
| BindingDB | 21393 |
| DrugBank | N/A |
Structure | 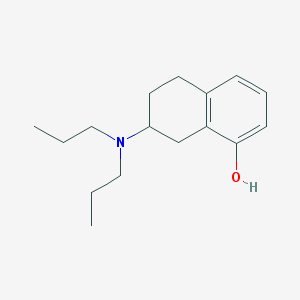 |
| Lipinski's druglikeness | This ligand satisfies Lipinski's rule of five. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| Basal | 12.98 (fM of GTP-gammaS bound) (mg of | PMID12213056 | ChEMBL |
| Basal | 13.5 (fM of GTP-gammaS bound) (mg of | PMID12213056 | ChEMBL |
| EC50 | 0.084 ug kg-1 | PMID11728188 | ChEMBL |
| EC50 | 0.137 ug kg-1 | PMID11728188 | ChEMBL |
| ED50 | 0.099 umol.kg-1 | PMID11101361 | ChEMBL |
| Hill coefficient | 1.01 - | PMID2521252 | ChEMBL |
| IC50 | 1.1 nM | PMID7699710, PMID8289207 | BindingDB,ChEMBL |
| IC50 | 2.1 nM | PMID8960552, PMID8759642 | BindingDB,ChEMBL |
| IC50 | 2.4 nM | PMID8863806, PMID7912735 | BindingDB,ChEMBL |
| IC50 | 3.0 nM | PMID3373482 | BindingDB |
| IC50 | 3.0 nM | PMID2537429 | BindingDB,ChEMBL |
| IC50 | 3.02 nM | PMID3373482 | ChEMBL |
| IC50 | 4.0 nM | PMID1433207 | BindingDB,ChEMBL |
| IC50 | 4.46 nM | PMID2571729 | ChEMBL |
| IC50 | 4.5 nM | PMID2571729 | BindingDB |
| IC50 | 8.06 nM | Bioorg. Med. Chem. Lett., (1997) 7:22:2857 | ChEMBL |
| IC50 | 8.1 nM | N/A | BindingDB |
| IC50 | 14.0 nM | PMID9083484 | BindingDB,ChEMBL |
| Increase | 91.85 % | PMID12213056 | ChEMBL |
| Kd | 2.5 nM | PMID7996545 | BindingDB,ChEMBL |
| Ki | 0.27 nM | Bioorg. Med. Chem. Lett., (1993) 3:10:2035 | ChEMBL |
| Ki | 0.27 nM | N/A | BindingDB |
| Ki | 0.47 nM | PMID10743959 | BindingDB |
| Ki | 0.47 nM | PMID10743959 | ChEMBL |
| Ki | 0.5 nM | PMID7861420, PMID7783152, PMID9083484 | BindingDB,ChEMBL |
| Ki | 0.5 nM | PMID7783152, PMID9083484 | BindingDB |
| Ki | 0.5 nM | PMID9083484 | BindingDB |
| Ki | 0.54 nM | N/A | BindingDB |
| Ki | 0.54 nM | Bioorg. Med. Chem. Lett., (1993) 3:10:2035 | ChEMBL |
| Ki | 0.63 nM | PMID8230102 | BindingDB,ChEMBL |
| Ki | 0.64 nM | PMID17900912 | BindingDB,ChEMBL |
| Ki | 0.8 nM | PMID19954866, PMID22133459 | BindingDB,ChEMBL |
| Ki | 0.8 nM | PMID26820556 | BindingDB |
| Ki | 1.0 nM | PMID2795604, PMID2140413 | BindingDB,ChEMBL |
| Ki | 1.02 nM | PMID24050112 | ChEMBL |
| Ki | 1.09 nM | Bioorg. Med. Chem. Lett., (1993) 3:10:2035 | ChEMBL |
| Ki | 1.1 nM | N/A | BindingDB |
| Ki | 1.18 nM | PMID7731013 | BindingDB,ChEMBL |
| Ki | 1.2 nM | PMID15588097, PMID14640559, PMID2965244, PMID12570387, PMID23466604, PMID2521252 | BindingDB,ChEMBL |
| Ki | 1.259 nM | PMID9046349 | ChEMBL |
| Ki | 1.4 nM | PMID7752194 | BindingDB |
| Ki | 1.413 nM | PMID9836623, PMID17300168, PMID17803293 | ChEMBL |
| Ki | 1.44 nM | PMID7752194, PMID1535661 | BindingDB,ChEMBL |
| Ki | 1.59 nM | PMID8289183 | ChEMBL |
| Ki | 1.6 nM | PMID8289183 | BindingDB |
| Ki | 1.862 nM | PMID8568799 | ChEMBL |
| Ki | 2.0 nM | PMID12877594, PMID12482417, PMID18760923, PMID3543362, PMID12166933 | BindingDB,ChEMBL |
| Ki | 2.1 nM | PMID11728188, PMID9836617, PMID2569041, PMID12477356 | BindingDB,ChEMBL |
| Ki | 2.26 nM | Bioorg. Med. Chem. Lett., (1993) 3:10:2035 | ChEMBL |
| Ki | 2.3 nM | N/A | BindingDB |
| Ki | 2.33 nM | PMID9888842 | BindingDB,ChEMBL |
| Ki | 2.8 nM | PMID8410989 | BindingDB,ChEMBL |
| Ki | 3.2 nM | PMID1995871 | BindingDB |
| Ki | 4.3 nM | PMID8568804, PMID8863803 | BindingDB,ChEMBL |
| Ki | 6.17 nM | PMID8584042 | BindingDB |
| Ki | 8.13 nM | PMID8584042 | BindingDB |
| Ki | 8.7 nM | PMID8340910, PMID8230131, PMID7932553 | BindingDB,ChEMBL |
| Ki | 27.0 nM | PMID8398139 | BindingDB |
| Ki | 975.0 nM | PMID2140413 | BindingDB,ChEMBL |
| KiAH | 1.16 nM | PMID1535661 | ChEMBL |
| Max | 23.83 (fM of GTP-gammaS bound) (mg of | PMID12213056 | ChEMBL |
| Max | 25.9 (fM of GTP-gammaS bound) (mg of | PMID12213056 | ChEMBL |
zhanglab![]() zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218