

You can:
| Name | Metabotropic glutamate receptor 5 |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | GRM5 |
| Synonym | mGlu5 receptor glutamate receptor GPRC1E mGluR5 |
| Disease | Central nervous system disease Chronic neuropathic pain Fragile X syndrome Major depressive disorder; GERD; Chronic neuropathic pain Autism [ Show all ] |
| Length | 1212 |
| Amino acid sequence | MVLLLILSVLLLKEDVRGSAQSSERRVVAHMPGDIIIGALFSVHHQPTVDKVHERKCGAVREQYGIQRVEAMLHTLERINSDPTLLPNITLGCEIRDSCWHSAVALEQSIEFIRDSLISSEEEEGLVRCVDGSSSSFRSKKPIVGVIGPGSSSVAIQVQNLLQLFNIPQIAYSATSMDLSDKTLFKYFMRVVPSDAQQARAMVDIVKRYNWTYVSAVHTEGNYGESGMEAFKDMSAKEGICIAHSYKIYSNAGEQSFDKLLKKLTSHLPKARVVACFCEGMTVRGLLMAMRRLGLAGEFLLLGSDGWADRYDVTDGYQREAVGGITIKLQSPDVKWFDDYYLKLRPETNHRNPWFQEFWQHRFQCRLEGFPQENSKYNKTCNSSLTLKTHHVQDSKMGFVINAIYSMAYGLHNMQMSLCPGYAGLCDAMKPIDGRKLLESLMKTNFTGVSGDTILFDENGDSPGRYEIMNFKEMGKDYFDYINVGSWDNGELKMDDDEVWSKKSNIIRSVCSEPCEKGQIKVIRKGEVSCCWTCTPCKENEYVFDEYTCKACQLGSWPTDDLTGCDLIPVQYLRWGDPEPIAAVVFACLGLLATLFVTVVFIIYRDTPVVKSSSRELCYIILAGICLGYLCTFCLIAKPKQIYCYLQRIGIGLSPAMSYSALVTKTNRIARILAGSKKKICTKKPRFMSACAQLVIAFILICIQLGIIVALFIMEPPDIMHDYPSIREVYLICNTTNLGVVTPLGYNGLLILSCTFYAFKTRNVPANFNEAKYIAFTMYTTCIIWLAFVPIYFGSNYKIITMCFSVSLSATVALGCMFVPKVYIILAKPERNVRSAFTTSTVVRMHVGDGKSSSAASRSSSLVNLWKRRGSSGETLRYKDRRLAQHKSEIECFTPKGSMGNGGRATMSSSNGKSVTWAQNEKSSRGQHLWQRLSIHINKKENPNQTAVIKPFPKSTESRGLGAGAGAGGSAGGVGATGGAGCAGAGPGGPESPDAGPKALYDVAEAEEHFPAPARPRSPSPISTLSHRAGSASRTDDDVPSLHSEPVARSSSSQGSLMEQISSVVTRFTANISELNSMMLSTAAPSPGVGAPLCSSYLIPKEIQLPTTMTTFAEIQPLPAIEVTGGAQPAAGAQAAGDAARESPAAGPEAAAAKPDLEELVALTPPSPFRDSVDSGSTTPNSPVSESALCIPSSPKYDTLIIRDYTQSSSSL |
| UniProt | P41594 |
| Protein Data Bank | 4oo9, 5cgc, 5cgd, 6ffh, 6ffi, 6n4x, 6n4y, 6n50, 6n51, 3lmk |
| GPCR-HGmod model | N/A |
| 3D structure model | This structure is from PDB ID 4oo9. |
| BioLiP | BL0176927,BL0176931, BL0176928,BL0176929,BL0176930, BL0281199, BL0322076, BL0322077, BL0407724, BL0438693, BL0438694, BL0438695,BL0438696,BL0438697, BL0407725, BL0438698,BL0438699 |
| Therapeutic Target Database | T99347 |
| ChEMBL | CHEMBL3227 |
| IUPHAR | 293 |
| DrugBank | BE0001192 |
| Name | CHEMBL363062 |
|---|---|
| Molecular formula | C22H16ClN3O |
| IUPAC name | 4-chloro-N-(2,5-diphenylpyrazol-3-yl)benzamide |
| Molecular weight | 373.84 |
| Hydrogen bond acceptor | 2 |
| Hydrogen bond donor | 1 |
| XlogP | 5.3 |
| Synonyms | BDBM50156077 4-Chloro-N-(2,5-diphenyl-2H-pyrazol-3-yl)-benzamide NSC-379650 AC1L7WJI 4-chloro-N-(2,5-diphenylpyrazol-3-yl)benzamide [ Show all ] |
| Inchi Key | ATESSTJCVXDGTE-UHFFFAOYSA-N |
| Inchi ID | InChI=1S/C22H16ClN3O/c23-18-13-11-17(12-14-18)22(27)24-21-15-20(16-7-3-1-4-8-16)25-26(21)19-9-5-2-6-10-19/h1-15H,(H,24,27) |
| PubChem CID | 342596 |
| ChEMBL | CHEMBL363062 |
| IUPHAR | N/A |
| BindingDB | 50156077 |
| DrugBank | N/A |
Structure | 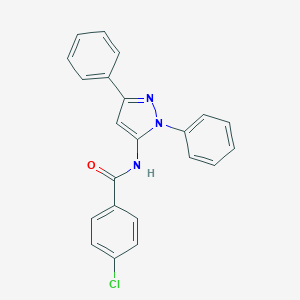 |
| Lipinski's druglikeness | This ligand has a partition coefficient log P greater than 5. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| EC50 | 20.0 nM | PMID15537338 | BindingDB,ChEMBL |
| Potentiation | 5.0 - | PMID15537338 | ChEMBL |
zhanglab![]() zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218