

You can:
| Name | NECA |
|---|---|
| Molecular formula | C12H16N6O4 |
| IUPAC name | (2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-N-ethyl-3,4-dihydroxyoxolane-2-carboxamide |
| Molecular weight | 308.298 |
| Hydrogen bond acceptor | 8 |
| Hydrogen bond donor | 4 |
| XlogP | -0.7 |
| Synonyms | (2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-N-ethyl-3,4-dihydroxyoxolane-2-carboxamide 35920-39-9 5'-Ethylcarboxamido Adenosine 84272-21-9 b-D-Ribofuranuronamide,1-(6-amino-9H-purin-9-yl)-1-deoxy-N-ethyl- [ Show all ] |
| Inchi Key | JADDQZYHOWSFJD-FLNNQWSLSA-N |
| Inchi ID | InChI=1S/C12H16N6O4/c1-2-14-11(21)8-6(19)7(20)12(22-8)18-4-17-5-9(13)15-3-16-10(5)18/h3-4,6-8,12,19-20H,2H2,1H3,(H,14,21)(H2,13,15,16)/t6-,7+,8-,12+/m0/s1 |
| PubChem CID | 448222 |
| ChEMBL | CHEMBL464859 |
| IUPHAR | 377, 425 |
| BindingDB | 21220 |
| DrugBank | N/A |
Structure | 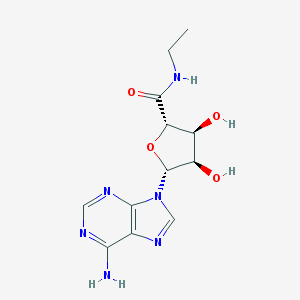 |
| Lipinski's druglikeness | This ligand satisfies Lipinski's rule of five. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| Activity | 0.75 pmol/ml | PMID20303771 | ChEMBL |
| Displacement | 0.0 % | PMID14584936 | ChEMBL |
| Displacement | 20.0 % | PMID14584936 | ChEMBL |
| EC50 | 12.58 nM | PMID25780876 | ChEMBL |
| EC50 | 12.59 nM | MedChemComm, (2014) 5:2:192 | ChEMBL |
| EC50 | 13.0 nM | PMID25780876 | BindingDB |
| EC50 | 17.38 nM | PMID17249651 | ChEMBL |
| EC50 | 17.6 nM | PMID24900277 | BindingDB,ChEMBL |
| EC50 | 18.0 nM | PMID23245803 | BindingDB,ChEMBL |
| EC50 | 21.9 nM | PMID17378544 | BindingDB,ChEMBL |
| EC50 | 40.0 nM | PMID11784146 | BindingDB,ChEMBL |
| EC50 | 45.0 nM | MedChemComm, (2015) 6:6:1178 | ChEMBL |
| EC50 | 50.0 nM | PMID15267242 | BindingDB,ChEMBL |
| EC50 | 251.19 nM | Med Chem Res, (2004) 13:1:88 | ChEMBL |
| EC50 | 338.84 nM | PMID26756468 | ChEMBL |
| EC50 | 339.0 nM | PMID26756468 | BindingDB |
| Emax | 100.0 % | PMID15267242 | ChEMBL |
| Emax | 102.0 % | PMID11784146 | ChEMBL |
| IC50 | 3.7 nM | PMID26988801, PMID27876250 | BindingDB |
| IC50 | 10.0 nM | PMID23582449 | ChEMBL |
| IC50 | 15.0 nM | PMID23466604 | ChEMBL |
| IC50 | 16.0 nM | PMID20031406 | BindingDB,ChEMBL |
| IC50 | 23.0 nM | PMID18588282 | ChEMBL |
| IC50 | 37.0 nM | PMID26988801, PMID27876250 | ChEMBL |
| IC50 | 64.0 nM | PMID18983139 | ChEMBL |
| IC50 | 67.0 nM | PMID20875743 | BindingDB,ChEMBL |
| Kd | 19.9526 nM | PMID9459566 | IUPHAR |
| Kdiss | 0.081 /s | PMID18045744 | ChEMBL |
| Ki | <10000.0 nM | PMID15734651 | BindingDB |
| Ki | 1.0 nM | PMID10714510 | BindingDB,ChEMBL |
| Ki | 1.5 nM | PMID14584936, PMID15734651 | BindingDB,ChEMBL |
| Ki | 1.99526 - 125.893 nM | PMID15476669, PMID15267242, PMID7775460, PMID9920286, PMID9179373, PMID14662005 | IUPHAR |
| Ki | 2.2 nM | PMID22104008, PMID15734651, PMID17378544 | BindingDB,ChEMBL |
| Ki | 5.2 nM | PMID9258366 | BindingDB |
| Ki | 5.24 nM | PMID9258366 | ChEMBL |
| Ki | 8.4 nM | PMID25780876 | BindingDB |
| Ki | 8.41 nM | PMID25780876 | ChEMBL |
| Ki | 9.3 nM | PMID15734651 | BindingDB |
| Ki | 12.0 nM | PMID23466604 | ChEMBL |
| Ki | 12.2 nM | PMID15481989 | BindingDB,ChEMBL |
| Ki | 12.4 nM | PMID17306548 | BindingDB,ChEMBL |
| Ki | 12.5 nM | PMID17228880 | BindingDB,ChEMBL |
| Ki | 15.0 nM | PMID11714602 | BindingDB |
| Ki | 15.1 nM | PMID11714602 | ChEMBL |
| Ki | 16.0 nM | PMID17927167, PMID15743197, PMID24077183, PMID18269230, PMID9572897, PMID11462973, PMID10956189, PMID16366607, PMID22257095 | BindingDB,ChEMBL |
| Ki | 19.1 nM | PMID14584936 | BindingDB,ChEMBL |
| Ki | 20.0 nM | PMID20408530, PMID23245803, PMID24900277, PMID10494877, PMID23200243, PMID24164628, PMID11459663, PMID22921089 | BindingDB,ChEMBL |
| Ki | 21.2 nM | PMID23200243 | ChEMBL |
| Ki | 21.4 nM | PMID14584936, PMID15734651 | BindingDB,ChEMBL |
| Ki | 21.5 nM | PMID9258366 | ChEMBL |
| Ki | 21.6 nM | PMID9258366 | ChEMBL |
| Ki | 22.0 nM | PMID9258366 | BindingDB |
| Ki | 24.6 nM | PMID14584936 | BindingDB,ChEMBL |
| Ki | 25.0 nM | PMID20031406 | BindingDB,ChEMBL |
| Ki | 29.2 nM | PMID14584936 | BindingDB,ChEMBL |
| Ki | 35.0 nM | PMID22486652 | BindingDB,ChEMBL |
| Ki | 52.0 nM | PMID18983139 | ChEMBL |
| Ki | 60.0 nM | PMID12672250, PMID15239649 | BindingDB,ChEMBL |
| Ki | 110.0 nM | PMID17967536 | BindingDB,ChEMBL |
| Ki | 124.0 nM | PMID16487705 | BindingDB,ChEMBL |
| Ki | 130.0 nM | PMID15267242 | BindingDB,ChEMBL |
| Ki | 680.0 nM | PMID17201410 | BindingDB,ChEMBL |
| koff | 0.03 min^-1 | DOI: http://dx.doi.org/10.6019/CHEMBL3885741 | ChEMBL |
| kon | 0.0005 nM^-1 min^-1 | DOI: http://dx.doi.org/10.6019/CHEMBL3885741 | ChEMBL |
| pKA | 5.9 - | PMID26756468 | ChEMBL |
| Potency ratio | 1.0 - | PMID7739005 | ChEMBL |
| Ratio | 1.0 - | PMID8863798, PMID8201607 | ChEMBL |
| Relative potency | 3.5 - | PMID12672250 | ChEMBL |
| T1/2 | 0.002367 hr | PMID18045744 | ChEMBL |
zhanglab![]() zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218