

You can:
| Name | Muscarinic acetylcholine receptor M1 |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | CHRM1 |
| Synonym | cholinergic receptor cholinergic receptor, muscarinic 1 cholinergic receptor, muscarinic 1, CNS Chrm-1 M1 receptor [ Show all ] |
| Disease | Functional bowel syndrome; Irritable bowel syndrome Glaucoma Peptic ulcer Parkinsonism; Extrapyramidal disorders secondary to neuroleptic drug therapy Visceral spasms [ Show all ] |
| Length | 460 |
| Amino acid sequence | MNTSAPPAVSPNITVLAPGKGPWQVAFIGITTGLLSLATVTGNLLVLISFKVNTELKTVNNYFLLSLACADLIIGTFSMNLYTTYLLMGHWALGTLACDLWLALDYVASNASVMNLLLISFDRYFSVTRPLSYRAKRTPRRAALMIGLAWLVSFVLWAPAILFWQYLVGERTVLAGQCYIQFLSQPIITFGTAMAAFYLPVTVMCTLYWRIYRETENRARELAALQGSETPGKGGGSSSSSERSQPGAEGSPETPPGRCCRCCRAPRLLQAYSWKEEEEEDEGSMESLTSSEGEEPGSEVVIKMPMVDPEAQAPTKQPPRSSPNTVKRPTKKGRDRAGKGQKPRGKEQLAKRKTFSLVKEKKAARTLSAILLAFILTWTPYNIMVLVSTFCKDCVPETLWELGYWLCYVNSTINPMCYALCNKAFRDTFRLLLLCRWDKRRWRKIPKRPGSVHRTPSRQC |
| UniProt | P11229 |
| Protein Data Bank | 5cxv |
| GPCR-HGmod model | P11229 |
| 3D structure model | This structure is from PDB ID 5cxv. |
| BioLiP | BL0339262, BL0339261, BL0339263 |
| Therapeutic Target Database | T28893 |
| ChEMBL | CHEMBL216 |
| IUPHAR | 13 |
| DrugBank | BE0000092 |
| Name | XANOMELINE |
|---|---|
| Molecular formula | C14H23N3OS |
| IUPAC name | 3-hexoxy-4-(1-methyl-3,6-dihydro-2H-pyridin-5-yl)-1,2,5-thiadiazole |
| Molecular weight | 281.418 |
| Hydrogen bond acceptor | 5 |
| Hydrogen bond donor | 0 |
| XlogP | 3.3 |
| Synonyms | L000694 Pyridine, 3-(4-(hexyloxy)-1,2,5-thiadiazol-3-yl)-1,2,5,6-tetrahydro-1-methyl- VP14766 (3-O-hexyloxy)-TZTP 5-(4-HEXYLOXY-[1,2,5]THIADIAZOL-3-YL)-1-METHYL-1,2,3,6-TETRAHYDRO-PYRIDINE [ Show all ] |
| Inchi Key | JOLJIIDDOBNFHW-UHFFFAOYSA-N |
| Inchi ID | InChI=1S/C14H23N3OS/c1-3-4-5-6-10-18-14-13(15-19-16-14)12-8-7-9-17(2)11-12/h8H,3-7,9-11H2,1-2H3 |
| PubChem CID | 60809 |
| ChEMBL | CHEMBL21536 |
| IUPHAR | 57 |
| BindingDB | 50003359 |
| DrugBank | N/A |
Structure | 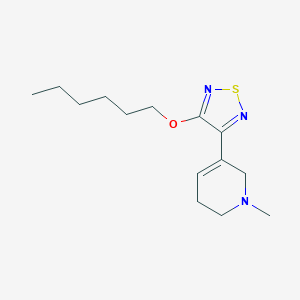 |
| Lipinski's druglikeness | This ligand satisfies Lipinski's rule of five. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| Activity | 770.0 % | PMID13678406 | ChEMBL |
| EC50 | 1.6 nM | PMID20684563 | BindingDB,ChEMBL |
| EC50 | 10.0 nM | PMID13678406 | ChEMBL |
| EC50 | 13.0 nM | PMID25275964 | BindingDB |
| EC50 | 13.18 nM | PMID25275964 | ChEMBL |
| EC50 | 15.85 nM | PMID25275964 | ChEMBL |
| EC50 | 16.0 nM | PMID25275964 | BindingDB |
| EC50 | 57.0 nM | PMID11741475 | BindingDB |
| EC50 | 57.0 nM | PMID11741475 | ChEMBL |
| EC50 | 67.0 nM | PMID26299349 | BindingDB |
| EC50 | 67.3 nM | PMID26299349 | ChEMBL |
| EC50 | 134.5 nM | PMID10354408 | ChEMBL |
| EC50 | 135.0 nM | PMID10354408 | BindingDB |
| EC50 | 158.49 nM | PMID17149881 | ChEMBL |
| EC50 | 200.0 nM | PMID9464368 | BindingDB,ChEMBL |
| EC50 | 316.23 nM | PMID17149881 | ChEMBL |
| EC50 | 2600.0 nM | PMID11741475 | BindingDB,ChEMBL |
| EC50 | 16218.0 nM | PMID25275964 | BindingDB |
| EC50 | 16218.1 nM | PMID25275964 | ChEMBL |
| Efficacy | 59.2 % | PMID26299349 | ChEMBL |
| Emax | 54.0 % | PMID25275964 | ChEMBL |
| Emax | 77.0 % | PMID25275964 | ChEMBL |
| Emax | 89.0 % | PMID20684563 | ChEMBL |
| Emax | 95.0 % | PMID25275964 | ChEMBL |
| IC50 | 0.008 nM | PMID1433209 | BindingDB,ChEMBL |
| IC50 | 18.0 nM | PMID17977730 | BindingDB |
| IC50 | 18.2 nM | PMID17977730 | ChEMBL |
| IC50 | 28.84 nM | PMID17977730 | ChEMBL |
| IC50 | 29.0 nM | PMID17977730 | BindingDB |
| IC50 | 81.0 nM | PMID17977730 | BindingDB |
| IC50 | 81.28 nM | PMID17977730 | ChEMBL |
| IC50 | 954.99 nM | PMID17977730 | ChEMBL |
| IC50 | 955.0 nM | PMID17977730 | BindingDB |
| IC50 | 2398.83 nM | PMID17977730 | ChEMBL |
| IC50 | 2399.0 nM | PMID17977730 | BindingDB |
| IC50 | 4897.79 nM | PMID17977730 | ChEMBL |
| IC50 | 4898.0 nM | PMID17977730 | BindingDB |
| Ki | 7.9 nM | PMID25275964 | BindingDB |
| Ki | 7.943 nM | PMID25275964, PMID18182302 | ChEMBL |
| Ki | 12.5893 - 199.526 nM | PMID9884068, PMID9614217, PMID10323594, PMID16675658 | IUPHAR |
| Ki | 42.0 nM | PMID10450949 | BindingDB,ChEMBL |
| Ki | 79.43 nM | PMID9884068 | BindingDB |
| Ki | 79.4328 nM | PMID9884068 | PDSP |
| Ki | 82.0 nM | PMID11741475 | BindingDB,ChEMBL |
| Ki | 158.49 nM | PMID17149881, PMID13678406 | ChEMBL |
| Ki | 2.51189e+14 nM | PMID17149881 | ChEMBL |
| logKd | 4.94 - | PMID20158205 | ChEMBL |
| logKd | 7.4 - | PMID20158205 | ChEMBL |
| Max | 65.6 % | PMID10354408 | ChEMBL |
| Max | 94.0 % | PMID1433209 | ChEMBL |
| Max PI | 63.0 % | PMID9873644 | ChEMBL |
| PI | 79.0 % | PMID10450949 | ChEMBL |
| PI metabolism | 47.0 % | PMID11741475 | ChEMBL |
| Smax | 31.0 % | PMID17149881 | ChEMBL |
| Smax | 71.0 % | PMID17149881 | ChEMBL |
| Smax | 86.0 % | PMID17149881 | ChEMBL |
| Smax | 150.0 % | PMID11741475 | ChEMBL |
| Smax | 180.0 % | PMID11741475 | ChEMBL |
| Stimulation | 70.0 % | Bioorg. Med. Chem. Lett., (1994) 4:18:2205 | ChEMBL |
zhanglab![]() zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218