

You can:
| Name | Beta-3 adrenergic receptor |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | ADRB3 |
| Synonym | ADRB Adrb-3 adrenergic receptor atypical beta-adrenoceptor beta 3-AR [ Show all ] |
| Disease | Urinary incontinence Diabetes Glaucoma Hypertension Irritable bowel syndrome [ Show all ] |
| Length | 408 |
| Amino acid sequence | MAPWPHENSSLAPWPDLPTLAPNTANTSGLPGVPWEAALAGALLALAVLATVGGNLLVIVAIAWTPRLQTMTNVFVTSLAAADLVMGLLVVPPAATLALTGHWPLGATGCELWTSVDVLCVTASIETLCALAVDRYLAVTNPLRYGALVTKRCARTAVVLVWVVSAAVSFAPIMSQWWRVGADAEAQRCHSNPRCCAFASNMPYVLLSSSVSFYLPLLVMLFVYARVFVVATRQLRLLRGELGRFPPEESPPAPSRSLAPAPVGTCAPPEGVPACGRRPARLLPLREHRALCTLGLIMGTFTLCWLPFFLANVLRALGGPSLVPGPAFLALNWLGYANSAFNPLIYCRSPDFRSAFRRLLCRCGRRLPPEPCAAARPALFPSGVPAARSSPAQPRLCQRLDGASWGVS |
| UniProt | P13945 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | P13945 |
| 3D structure model | This predicted structure model is from GPCR-EXP P13945. |
| BioLiP | N/A |
| Therapeutic Target Database | T51408 |
| ChEMBL | CHEMBL246 |
| IUPHAR | 30 |
| DrugBank | BE0001012, BE0004872 |
| Name | UNII-P50A5YTO0O |
|---|---|
| Molecular formula | C31H27F3N4O3S2 |
| IUPAC name | N-[4-[2-[[(2R)-2-hydroxy-2-pyridin-3-ylethyl]amino]ethyl]phenyl]-4-[4-[4-(trifluoromethyl)phenyl]-1,3-thiazol-2-yl]benzenesulfonamide |
| Molecular weight | 624.697 |
| Hydrogen bond acceptor | 11 |
| Hydrogen bond donor | 3 |
| XlogP | 5.3 |
| Synonyms | (R)-N-(4-(2-(2-hydroxy-2-(pyridin-3-yl)ethylamino)ethyl)phenyl)-4-(4-(4-(trifluoromethyl)phenyl)thiazol-2-yl)benzenesulfonamide CHEMBL111201 SCHEMBL1276546 N-[4-[2-[[(2R)-2-hydroxy-2-pyridin-3-ylethyl]amino]ethyl]phenyl]-4-[4-[4-(trifluoromethyl)phenyl]-1,3-thiazol-2-yl]benzenesulfonamide (1R)-1-(3-Pyridyl)-2-[4-[[4-[4-[4-(trifluoromethyl)phenyl]-2-thiazolyl]phenyl]sulfonylamino]phenethylamino]ethanol [ Show all ] |
| Inchi Key | MSOUIIHPMJCUNI-LJAQVGFWSA-N |
| Inchi ID | InChI=1S/C31H27F3N4O3S2/c32-31(33,34)25-9-5-22(6-10-25)28-20-42-30(37-28)23-7-13-27(14-8-23)43(40,41)38-26-11-3-21(4-12-26)15-17-36-19-29(39)24-2-1-16-35-18-24/h1-14,16,18,20,29,36,38-39H,15,17,19H2/t29-/m0/s1 |
| PubChem CID | 3038500 |
| ChEMBL | CHEMBL111201 |
| IUPHAR | N/A |
| BindingDB | 50092645 |
| DrugBank | N/A |
Structure | 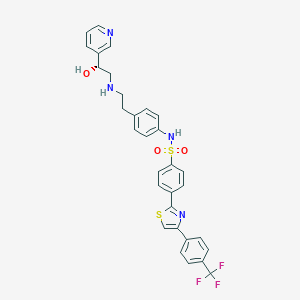 |
| Lipinski's druglikeness | This ligand has more than 10 hydrogen bond acceptor. This ligand is heavier than 500 daltons. This ligand has a partition coefficient log P greater than 5. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| ACT | 94.0 % | PMID11052788 | ChEMBL |
| Activity | 94.0 % | PMID20181479 | ChEMBL |
| EC50 | 2.0 nM | PMID26709102 | BindingDB |
| EC50 | 2.0 nM | PMID26709102 | ChEMBL |
| EC50 | 3.0 nM | PMID17632003, PMID15149682 | BindingDB,ChEMBL |
| EC50 | 3.6 nM | PMID11052788, PMID20181479 | BindingDB,ChEMBL |
| EC50 | 7.9 nM | PMID26709102 | BindingDB,ChEMBL |
| IA | 92.0 - | PMID15149682 | ChEMBL |
| IC50 | 46.0 nM | PMID11052788 | BindingDB,ChEMBL |
| Intrinsic activity | 92.0 % | PMID17632003 | ChEMBL |
| max activation | 93.0 % | PMID26709102 | ChEMBL |
| max activation | 106.0 % | PMID26709102 | ChEMBL |
zhanglab![]() zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218