

You can:
| Name | Neuromedin-K receptor |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | TACR3 |
| Synonym | SP-N receptor Tac3r Nmkr NKR NK3 receptor [ Show all ] |
| Disease | Schizophrenia Schizophrenia; Schizoaffective disorders Psychotic disorders Psychiatric disorder Irritable bowel syndrome [ Show all ] |
| Length | 465 |
| Amino acid sequence | MATLPAAETWIDGGGGVGADAVNLTASLAAGAATGAVETGWLQLLDQAGNLSSSPSALGLPVASPAPSQPWANLTNQFVQPSWRIALWSLAYGVVVAVAVLGNLIVIWIILAHKRMRTVTNYFLVNLAFSDASMAAFNTLVNFIYALHSEWYFGANYCRFQNFFPITAVFASIYSMTAIAVDRYMAIIDPLKPRLSATATKIVIGSIWILAFLLAFPQCLYSKTKVMPGRTLCFVQWPEGPKQHFTYHIIVIILVYCFPLLIMGITYTIVGITLWGGEIPGDTCDKYHEQLKAKRKVVKMMIIVVMTFAICWLPYHIYFILTAIYQQLNRWKYIQQVYLASFWLAMSSTMYNPIIYCCLNKRFRAGFKRAFRWCPFIKVSSYDELELKTTRFHPNRQSSMYTVTRMESMTVVFDPNDADTTRSSRKKRATPRDPSFNGCSRRNSKSASATSSFISSPYTSVDEYS |
| UniProt | P29371 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | P29371 |
| 3D structure model | This predicted structure model is from GPCR-EXP P29371. |
| BioLiP | N/A |
| Therapeutic Target Database | T29683 |
| ChEMBL | CHEMBL4429 |
| IUPHAR | 362 |
| DrugBank | BE0002371 |
| Name | Talnetant |
|---|---|
| Molecular formula | C25H22N2O2 |
| IUPAC name | 3-hydroxy-2-phenyl-N-[(1S)-1-phenylpropyl]quinoline-4-carboxamide |
| Molecular weight | 382.463 |
| Hydrogen bond acceptor | 3 |
| Hydrogen bond donor | 2 |
| XlogP | 5.7 |
| Synonyms | (S)-(-)-N-(R-ethylbenzyl)-3-hydroxy-2-phenylquinoline-4-carboxamide 3-Hydroxy-2-phenyl-quinoline-4-carboxylic acid ((S)-1-phenyl-propyl)-amide AKOS024458283 CZ3T9T146K N-((S)-alpha-Ethylbenzyl)-3-hydroxy-2-phenylcinchoninamide [ Show all ] |
| Inchi Key | BIAVGWDGIJKWRM-FQEVSTJZSA-N |
| Inchi ID | InChI=1S/C25H22N2O2/c1-2-20(17-11-5-3-6-12-17)27-25(29)22-19-15-9-10-16-21(19)26-23(24(22)28)18-13-7-4-8-14-18/h3-16,20,28H,2H2,1H3,(H,27,29)/t20-/m0/s1 |
| PubChem CID | 5311424 |
| ChEMBL | CHEMBL10188 |
| IUPHAR | 2132 |
| BindingDB | 50051293 |
| DrugBank | N/A |
Structure | 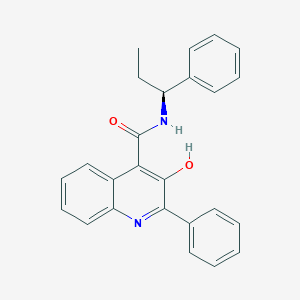 |
| Lipinski's druglikeness | This ligand has a partition coefficient log P greater than 5. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| IC50 | 2.4 nM | PMID16950620 | BindingDB,ChEMBL |
| IC50 | 6.1 nM | PMID21047106 | BindingDB,ChEMBL |
| IC50 | 8.4 nM | PMID27298001 | BindingDB,ChEMBL |
| IC50 | 16.6 nM | PMID21047106 | BindingDB,ChEMBL |
| Kb | 3.0 nM | PMID8691422 | ChEMBL |
| Kd | 5.75 nM | PMID19817444, PMID22574973 | BindingDB |
| Kd | 5.754 nM | PMID19817444, PMID22574973 | ChEMBL |
| Ki | 1.0 nM | PMID9190866, PMID8691422 | BindingDB,ChEMBL |
| Ki | 1.0 - 39.8107 nM | PMID9190866, PMID15265501, PMID11226387, PMID8691422 | IUPHAR |
| Ki | 1.4 nM | PMID11356103 | BindingDB,ChEMBL |
| Ki | 1.995 nM | PMID24374277 | ChEMBL |
| Ki | 2.0 nM | PMID24374277 | BindingDB |
| Ki | 2.51 nM | PMID21047106 | BindingDB |
| Ki | 3.0 nM | PMID19817444 | BindingDB,ChEMBL |
| Ki | 3.6 nM | PMID22574973 | BindingDB,ChEMBL |
| Ki | 7.4 nM | PMID21376585 | BindingDB,ChEMBL |
| Ki | 7.9 nM | PMID25738882 | BindingDB |
| Ki | 7.943 nM | PMID25738882 | ChEMBL |
| Ki | 254.0 nM | PMID10090788 | BindingDB,ChEMBL |
| Occ90 | 75.0 mg kg-1 | PMID16950620 | ChEMBL |
zhanglab![]() zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218