

You can:
| Name | Histamine H4 receptor |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | HRH4 |
| Synonym | Pfi-013 SP9144 HH4R H4R H4 receptor [ Show all ] |
| Disease | Allergic rhinitis Asthma Inflammatory disease Rheumatoid arthritis |
| Length | 390 |
| Amino acid sequence | MPDTNSTINLSLSTRVTLAFFMSLVAFAIMLGNALVILAFVVDKNLRHRSSYFFLNLAISDFFVGVISIPLYIPHTLFEWDFGKEICVFWLTTDYLLCTASVYNIVLISYDRYLSVSNAVSYRTQHTGVLKIVTLMVAVWVLAFLVNGPMILVSESWKDEGSECEPGFFSEWYILAITSFLEFVIPVILVAYFNMNIYWSLWKRDHLSRCQSHPGLTAVSSNICGHSFRGRLSSRRSLSASTEVPASFHSERQRRKSSLMFSSRTKMNSNTIASKMGSFSQSDSVALHQREHVELLRARRLAKSLAILLGVFAVCWAPYSLFTIVLSFYSSATGPKSVWYRIAFWLQWFNSFVNPLLYPLCHKRFQKAFLKIFCIKKQPLPSQHSRSVSS |
| UniProt | Q9H3N8 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | Q9H3N8 |
| 3D structure model | This predicted structure model is from GPCR-EXP Q9H3N8. |
| BioLiP | N/A |
| Therapeutic Target Database | T26500 |
| ChEMBL | CHEMBL3759 |
| IUPHAR | 265 |
| DrugBank | BE0000146 |
| Name | imetit |
|---|---|
| Molecular formula | C6H10N4S |
| IUPAC name | 2-(1H-imidazol-5-yl)ethyl carbamimidothioate |
| Molecular weight | 170.234 |
| Hydrogen bond acceptor | 3 |
| Hydrogen bond donor | 3 |
| XlogP | 0.2 |
| Synonyms | NCGC00015536-05 UNII-677MJ4VPZC 2-(1H-Imidazol-4-yl)ethyl carbamimidothioate AKOS006273468 CHEBI:64156 [ Show all ] |
| Inchi Key | PEHSVUKQDJULKE-UHFFFAOYSA-N |
| Inchi ID | InChI=1S/C6H10N4S/c7-6(8)11-2-1-5-3-9-4-10-5/h3-4H,1-2H2,(H3,7,8)(H,9,10) |
| PubChem CID | 3692 |
| ChEMBL | CHEMBL19439 |
| IUPHAR | 1250 |
| BindingDB | 22911 |
| DrugBank | N/A |
Structure | 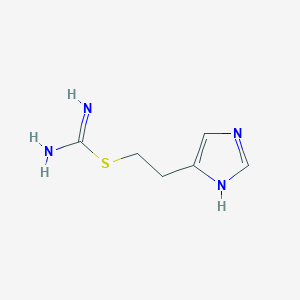 |
| Lipinski's druglikeness | This ligand satisfies Lipinski's rule of five. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| EC50 | 12.59 nM | PMID21044842, PMID21062081 | BindingDB,ChEMBL |
| EC50 | 51.0 nM | PMID12723960 | BindingDB,ChEMBL |
| Intrinsic activity | 0.9 - | PMID21044842 | ChEMBL |
| Ki | 1.25893 - 6.30957 nM | PMID11179434, PMID11561071, PMID11181941, PMID11179436 | IUPHAR |
| Ki | 1.6 nM | PMID21498080 | BindingDB,ChEMBL |
| Ki | 2.7 nM | PMID11179434 | PDSP,BindingDB |
| Ki | 3.6 nM | PMID18459760 | BindingDB,ChEMBL |
| Ki | 6.0 nM | PMID15947036 | BindingDB |
| Ki | 6.31 nM | PMID19414267, PMID21044842, PMID22003888 | BindingDB,ChEMBL |
| Ki | 3795.6 nM | PMID11179435 | PDSP,BindingDB |
zhanglab![]() zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218