

You can:
| Name | Alpha-2A adrenergic receptor |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | ADRA2A |
| Synonym | alpha2D alpha2A-AR alpha2A-adrenoceptor alpha2A-adrenergic receptor alpha2A [ Show all ] |
| Disease | Attention deficit hyperactivity disorder Sexual dysfunction Pain |
| Length | 450 |
| Amino acid sequence | MGSLQPDAGNASWNGTEAPGGGARATPYSLQVTLTLVCLAGLLMLLTVFGNVLVIIAVFTSRALKAPQNLFLVSLASADILVATLVIPFSLANEVMGYWYFGKAWCEIYLALDVLFCTSSIVHLCAISLDRYWSITQAIEYNLKRTPRRIKAIIITVWVISAVISFPPLISIEKKGGGGGPQPAEPRCEINDQKWYVISSCIGSFFAPCLIMILVYVRIYQIAKRRTRVPPSRRGPDAVAAPPGGTERRPNGLGPERSAGPGGAEAEPLPTQLNGAPGEPAPAGPRDTDALDLEESSSSDHAERPPGPRRPERGPRGKGKARASQVKPGDSLPRRGPGATGIGTPAAGPGEERVGAAKASRWRGRQNREKRFTFVLAVVIGVFVVCWFPFFFTYTLTAVGCSVPRTLFKFFFWFGYCNSSLNPVIYTIFNHDFRRAFKKILCRGDRKRIV |
| UniProt | P08913 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | P08913 |
| 3D structure model | This predicted structure model is from GPCR-EXP P08913. |
| BioLiP | N/A |
| Therapeutic Target Database | T11448 |
| ChEMBL | CHEMBL1867 |
| IUPHAR | 25 |
| DrugBank | BE0000289 |
| Name | Yohimbine |
|---|---|
| Molecular formula | C21H26N2O3 |
| IUPAC name | methyl (1S,15R,18S,19R,20S)-18-hydroxy-1,3,11,12,14,15,16,17,18,19,20,21-dodecahydroyohimban-19-carboxylate |
| Molecular weight | 354.45 |
| Hydrogen bond acceptor | 4 |
| Hydrogen bond donor | 2 |
| XlogP | 2.9 |
| Synonyms | NCGC00025018-07 Baron-X Quebrachin Bio2_000458 SR-01000075297-5 [ Show all ] |
| Inchi Key | BLGXFZZNTVWLAY-SCYLSFHTSA-N |
| Inchi ID | InChI=1S/C21H26N2O3/c1-26-21(25)19-15-10-17-20-14(13-4-2-3-5-16(13)22-20)8-9-23(17)11-12(15)6-7-18(19)24/h2-5,12,15,17-19,22,24H,6-11H2,1H3/t12-,15-,17-,18-,19+/m0/s1 |
| PubChem CID | 8969 |
| ChEMBL | CHEMBL15245 |
| IUPHAR | 102 |
| BindingDB | 50013515, 50203564 |
| DrugBank | DB01392 |
Structure | 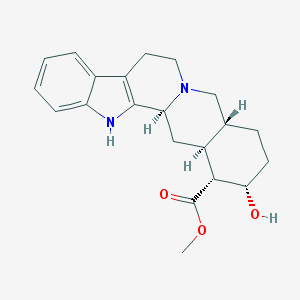 |
| Lipinski's druglikeness | This ligand satisfies Lipinski's rule of five. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| N/A | N/A | DrugBank | |
| EC50 | 8.5 nM | PMID16562853 | BindingDB,ChEMBL |
| IC50 | 3.67 nM | PMID19788200 | BindingDB |
| IC50 | 6.151 nM | DrugMatrix in vitro pharmacology data | ChEMBL |
| IC50 | 8.4 nM | PMID23403082 | ChEMBL |
| Inhibition | -5.0 % | PMID23403082 | ChEMBL |
| Ki | 0.32 nM | PMID1353247 | BindingDB |
| Ki | 0.42 nM | PMID10762040 | ChEMBL |
| Ki | 0.42 nM | PMID10762040 | BindingDB |
| Ki | 0.43 nM | PMID7908054 | BindingDB |
| Ki | 0.44 nM | PMID1353247, PMID1656026 | BindingDB |
| Ki | 0.5 nM | PMID1353247 | BindingDB |
| Ki | 0.61 nM | PMID7908054 | BindingDB |
| Ki | 0.630958 - 3.98108 nM | PMID7996470, PMID1353247, PMID7908642 | IUPHAR |
| Ki | 0.65 nM | PMID1353247, PMID1656026 | BindingDB |
| Ki | 0.71 nM | PMID9400006 | BindingDB |
| Ki | 1.2 nM | PMID9605427 | BindingDB |
| Ki | 1.4 nM | PMID15911252 | ChEMBL |
| Ki | 1.6 nM | PMID11408545, PMID6149136 | BindingDB |
| Ki | 1.8 nM | PMID17257841 | BindingDB |
| Ki | 2.307 nM | DrugMatrix in vitro pharmacology data | ChEMBL |
| Ki | 2.5 nM | PMID11132243 | BindingDB |
| Ki | 2.7 nM | PMID6149136 | BindingDB |
| Ki | 3.1 nM | PMID23403082, PMID10602703 | BindingDB,ChEMBL |
| Ki | 3.16 nM | PMID10611634 | BindingDB |
| Ki | 3.5 nM | PMID11408545 | BindingDB |
| Ki | 3.6 nM | PMID7855217 | BindingDB |
| Ki | 3.71 nM | PMID7996470 | BindingDB |
| Ki | 3.76 nM | PMID7996470 | BindingDB |
| Ki | 6.3 nM | PMID10611634 | BindingDB |
| Ki | 7.5 nM | PMID7562902 | BindingDB,ChEMBL |
| Ki | 7.94 nM | PMID10611634 | BindingDB |
| Ki | 132.0 nM | PMID1656026 | BindingDB |
zhanglab![]() zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218