

You can:
| Name | Kappa-type opioid receptor |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | OPRK1 |
| Synonym | K-OR-1 KOPr OP2 KOP KOR-1 [ Show all ] |
| Disease | Obesity Opiate dependence Inflammatory bowel disease Erythema Diarrhea-predominant IBS [ Show all ] |
| Length | 380 |
| Amino acid sequence | MDSPIQIFRGEPGPTCAPSACLPPNSSAWFPGWAEPDSNGSAGSEDAQLEPAHISPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTTTMPFQSTVYLMNSWPFGDVLCKIVISIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVREDVDVIECSLQFPDDDYSWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSGSREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSTSHSTAALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFPLKMRMERQSTSRVRNTVQDPAYLRDIDGMNKPV |
| UniProt | P41145 |
| Protein Data Bank | 6b73, 4djh |
| GPCR-HGmod model | P41145 |
| 3D structure model | This structure is from PDB ID 6b73. |
| BioLiP | BL0402244,BL0402246, BL0224693,BL0224694, BL0402243,BL0402245 |
| Therapeutic Target Database | T60693 |
| ChEMBL | CHEMBL237 |
| IUPHAR | 318 |
| DrugBank | BE0000632 |
| Name | morphine |
|---|---|
| Molecular formula | C17H19NO3 |
| IUPAC name | (4R,4aR,7S,7aR,12bS)-3-methyl-2,4,4a,7,7a,13-hexahydro-1H-4,12-methanobenzofuro[3,2-e]isoquinoline-7,9-diol |
| Molecular weight | 285.343 |
| Hydrogen bond acceptor | 4 |
| Hydrogen bond donor | 2 |
| XlogP | 0.8 |
| Synonyms | UNII-76I7G6D29C 4-methyl-(1S,5R,13R,14S,17R)-12-oxa-4-azapentacyclo[9.6.1.01,13.05,17.07,18]octadeca-7(18),8,10,15-tetraene-10,14-diol ;sulphate salt(morphine) Morphina 4-methyl-12-oxa-4-azapentacyclo[9.6.1.01,13.05,17.07,18]octadeca-7(18),8,10,15-tetraene-10,14-diol (morphine) Morphine 0.1 mg/ml in Methanol [ Show all ] |
| Inchi Key | BQJCRHHNABKAKU-KBQPJGBKSA-N |
| Inchi ID | InChI=1S/C17H19NO3/c1-18-7-6-17-10-3-5-13(20)16(17)21-15-12(19)4-2-9(14(15)17)8-11(10)18/h2-5,10-11,13,16,19-20H,6-8H2,1H3/t10-,11+,13-,16-,17-/m0/s1 |
| PubChem CID | 5288826 |
| ChEMBL | CHEMBL70 |
| IUPHAR | 1627 |
| BindingDB | 50000092 |
| DrugBank | DB00295 |
Structure | 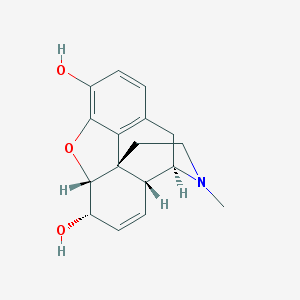 |
| Lipinski's druglikeness | This ligand satisfies Lipinski's rule of five. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| N/A | N/A | DrugBank | |
| Activity | 31.0 - | PMID7515442 | ChEMBL |
| Activity | 62.0 % | PMID21235243, PMID16913723, PMID19253983, PMID17887741 | ChEMBL |
| EC50 | 400.0 nM | PMID23880538 | ChEMBL |
| EC50 | 484.0 nM | PMID16913723, PMID21235243, PMID19253983, PMID17887741 | BindingDB,ChEMBL |
| ED50 | 1140.0 nM | PMID20055417 | ChEMBL |
| Emax | 28.0 % | PMID20055417 | ChEMBL |
| IC50 | 217.0 nM | PMID11585443 | BindingDB |
| IC50 | 1705.0 nM | PMID12502358 | BindingDB |
| IC50 | 1705.1 nM | PMID12502358 | ChEMBL |
| Inhibition | 90.0 % | PMID23880538 | ChEMBL |
| Ki | 6.9 nM | PMID19932964 | BindingDB,ChEMBL |
| Ki | 11.1 nM | PMID7932177 | BindingDB |
| Ki | 13.0 nM | PMID22680612 | BindingDB |
| Ki | 13.3 nM | PMID22680612 | ChEMBL |
| Ki | 24.0 nM | PMID22439881, PMID16392810, PMID15055988 | BindingDB,ChEMBL |
| Ki | 33.0 nM | PMID1315868 | BindingDB,ChEMBL |
| Ki | 33.7 nM | PMID20478711 | BindingDB,ChEMBL |
| Ki | 46.9 nM | PMID16913723, PMID17887741, PMID9686407 | BindingDB,ChEMBL |
| Ki | 47.0 nM | PMID16913723, PMID17887741 | BindingDB |
| Ki | 50.1187 nM | PMID9686407 | IUPHAR |
| Ki | 52.0 nM | PMID12372519 | BindingDB |
| Ki | 52.14 nM | PMID12372519 | ChEMBL |
| Ki | 60.9 nM | PMID23419026 | ChEMBL |
| Ki | 61.0 nM | PMID23419026 | BindingDB |
| Ki | 64.0 nM | PMID12930147 | BindingDB,ChEMBL |
| Ki | 65.5 nM | PMID18311899 | ChEMBL |
| Ki | 86.0 nM | PMID2567782 | BindingDB,ChEMBL |
| Ki | 113.0 nM | PMID12699394 | BindingDB,ChEMBL |
| Ki | 120.0 nM | PMID26390077 | BindingDB,ChEMBL |
| Ki | 260.0 nM | PMID12166947, PMID9651168 | BindingDB,ChEMBL |
| Ki | 299.0 nM | PMID25599950, PMID23618710, PMID22341895, PMID23880358 | BindingDB,ChEMBL |
| Ki | 538.0 nM | PMID8114680 | BindingDB |
| Ki | 1900.0 nM | , Bioorg. Med. Chem. Lett., (1992) 2:7:715 | BindingDB,ChEMBL |
zhanglab![]() zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218