

You can:
| Name | Glucagon-like peptide 1 receptor |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | GLP1R |
| Synonym | glucagon-like peptide 1 receptor GLP-1R GLP-1-R GLP-1 receptor |
| Disease | Type 1/2 diabetes Type 1 diabetes Obesity Non-insulin dependent diabetes Non-alcoholic steatohepatitis [ Show all ] |
| Length | 463 |
| Amino acid sequence | MAGAPGPLRLALLLLGMVGRAGPRPQGATVSLWETVQKWREYRRQCQRSLTEDPPPATDLFCNRTFDEYACWPDGEPGSFVNVSCPWYLPWASSVPQGHVYRFCTAEGLWLQKDNSSLPWRDLSECEESKRGERSSPEEQLLFLYIIYTVGYALSFSALVIASAILLGFRHLHCTRNYIHLNLFASFILRALSVFIKDAALKWMYSTAAQQHQWDGLLSYQDSLSCRLVFLLMQYCVAANYYWLLVEGVYLYTLLAFSVLSEQWIFRLYVSIGWGVPLLFVVPWGIVKYLYEDEGCWTRNSNMNYWLIIRLPILFAIGVNFLIFVRVICIVVSKLKANLMCKTDIKCRLAKSTLTLIPLLGTHEVIFAFVMDEHARGTLRFIKLFTELSFTSFQGLMVAILYCFVNNEVQLEFRKSWERWRLEHLHIQRDSSMKPLKCPTSSLSSGATAGSSMYTATCQASCS |
| UniProt | P43220 |
| Protein Data Bank | 5vex, 3c59, 3c5t, 5nx2, 3iol, 4zgm, 5otu, 5vew, 5otw, 5otx, 5otv |
| GPCR-HGmod model | P43220 |
| 3D structure model | This structure is from PDB ID 5vex. |
| BioLiP | BL0418498,BL0418499, BL0143794, BL0143795, BL0167479, BL0167480, BL0324354, BL0324355,BL0324356, BL0378791,BL0378792, BL0379513,BL0379514, BL0418500,BL0418501, BL0380967, BL0418494,BL0418495, BL0418496,BL0418497, BL0143732, BL0143731, BL0380966 |
| Therapeutic Target Database | T36075 |
| ChEMBL | CHEMBL1784 |
| IUPHAR | 249 |
| DrugBank | BE0000857 |
| Name | MLS000544661 |
|---|---|
| Molecular formula | C18H12Br2N4 |
| IUPAC name | 2,4-bis(3-bromophenyl)-7-methylpyrazolo[3,4-d]pyridazine |
| Molecular weight | 444.13 |
| Hydrogen bond acceptor | 3 |
| Hydrogen bond donor | 0 |
| XlogP | 4.6 |
| Synonyms | 2,4-bis(3-bromophenyl)-7-methyl-2H-pyrazolo[3,4-d]pyridazine CHEMBL1536422 MolPort-002-817-998 BDBM88589 cid_1214669 [ Show all ] |
| Inchi Key | AECFCCDRHPPLMX-UHFFFAOYSA-N |
| Inchi ID | InChI=1S/C18H12Br2N4/c1-11-17-16(10-24(23-17)15-7-3-6-14(20)9-15)18(22-21-11)12-4-2-5-13(19)8-12/h2-10H,1H3 |
| PubChem CID | 1214669 |
| ChEMBL | CHEMBL1536422 |
| IUPHAR | N/A |
| BindingDB | 88589 |
| DrugBank | N/A |
Structure | 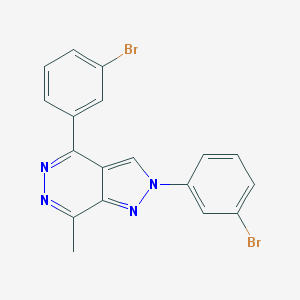 |
| Lipinski's druglikeness | This ligand satisfies Lipinski's rule of five. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| Potency | 7943.3 nM | PubChem BioAssay data set | ChEMBL |
zhanglab![]() zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218