

You can:
| Name | Adenosine receptor A3 |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | ADORA3 |
| Synonym | ARA3 Adenosine receptor A3 A3AR A3 receptor TGPCR1 |
| Disease | Cerebrovascular ischaemia Malaria Ischemia Inflammation Hepatocellular carcinoma; Hepatitis C virus infection [ Show all ] |
| Length | 318 |
| Amino acid sequence | MPNNSTALSLANVTYITMEIFIGLCAIVGNVLVICVVKLNPSLQTTTFYFIVSLALADIAVGVLVMPLAIVVSLGITIHFYSCLFMTCLLLIFTHASIMSLLAIAVDRYLRVKLTVRYKRVTTHRRIWLALGLCWLVSFLVGLTPMFGWNMKLTSEYHRNVTFLSCQFVSVMRMDYMVYFSFLTWIFIPLVVMCAIYLDIFYIIRNKLSLNLSNSKETGAFYGREFKTAKSLFLVLFLFALSWLPLSIINCIIYFNGEVPQLVLYMGILLSHANSMMNPIVYAYKIKKFKETYLLILKACVVCHPSDSLDTSIEKNSE |
| UniProt | P0DMS8 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | N/A |
| 3D structure model | No available structures or models |
| BioLiP | N/A |
| Therapeutic Target Database | T36059 |
| ChEMBL | CHEMBL256 |
| IUPHAR | 21 |
| DrugBank | BE0000354 |
| Name | Sch 58261 |
|---|---|
| Molecular formula | C18H15N7O |
| IUPAC name | 4-(furan-2-yl)-10-(2-phenylethyl)-3,5,6,8,10,11-hexazatricyclo[7.3.0.02,6]dodeca-1(9),2,4,7,11-pentaen-7-amine |
| Molecular weight | 345.366 |
| Hydrogen bond acceptor | 6 |
| Hydrogen bond donor | 1 |
| XlogP | 2.4 |
| Synonyms | UNII-4309023MAH 4-(furan-2-yl)-10-(2-phenylethyl)-3,5,6,8,10,11-hexaazatricyclo[7.3.0.0^{2,6}]dodeca-1(9),2,4,7,11-pentaen-7-amine 7H-Pyrazolo(4,3-e)(1,2,4)triazolo(1,5-c)pyrimidin-5-amine, 2-(2-furanyl)-7-(2-phenylethyl)- B6995 DTXSID80166799 [ Show all ] |
| Inchi Key | UTLPKQYUXOEJIL-UHFFFAOYSA-N |
| Inchi ID | InChI=1S/C18H15N7O/c19-18-22-16-13(11-20-24(16)9-8-12-5-2-1-3-6-12)17-21-15(23-25(17)18)14-7-4-10-26-14/h1-7,10-11H,8-9H2,(H2,19,22) |
| PubChem CID | 176408 |
| ChEMBL | CHEMBL17127 |
| IUPHAR | 431, 403 |
| BindingDB | 50048466 |
| DrugBank | N/A |
Structure | 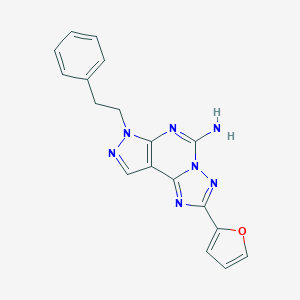 |
| Lipinski's druglikeness | This ligand satisfies Lipinski's rule of five. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| Ki | <1000.0 nM | PMID12646033 | BindingDB,ChEMBL |
| Ki | <10000.0 nM | PMID19501513, PMID9933143, PMID22204739, PMID9622554, PMID11809867, PMID10779381, PMID11754583 | PDSP,BindingDB,ChEMBL |
| Ki | 1200.0 nM | PMID24164628, PMID16518376 | BindingDB,IUPHAR,ChEMBL |
zhanglab![]() zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218