

You can:
| Name | C-C chemokine receptor type 2 |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | CCR2 |
| Synonym | MCP-1-R Monocyte chemoattractant protein 1 receptor CKR2 MCP-1 receptor chemokine receptor CCR2 [ Show all ] |
| Disease | Chronic obstructive pulmonary disease Neuropathic pain Multiple scierosis Metastatic cancer; Multiple scierosis Inflammatory disease [ Show all ] |
| Length | 374 |
| Amino acid sequence | MLSTSRSRFIRNTNESGEEVTTFFDYDYGAPCHKFDVKQIGAQLLPPLYSLVFIFGFVGNMLVVLILINCKKLKCLTDIYLLNLAISDLLFLITLPLWAHSAANEWVFGNAMCKLFTGLYHIGYFGGIFFIILLTIDRYLAIVHAVFALKARTVTFGVVTSVITWLVAVFASVPGIIFTKCQKEDSVYVCGPYFPRGWNNFHTIMRNILGLVLPLLIMVICYSGILKTLLRCRNEKKRHRAVRVIFTIMIVYFLFWTPYNIVILLNTFQEFFGLSNCESTSQLDQATQVTETLGMTHCCINPIIYAFVGEKFRSLFHIALGCRIAPLQKPVCGGPGVRPGKNVKVTTQGLLDGRGKGKSIGRAPEASLQDKEGA |
| UniProt | P41597 |
| Protein Data Bank | 6gpx, 6gps |
| GPCR-HGmod model | P41597 |
| 3D structure model | This structure is from PDB ID 6gpx. |
| BioLiP | BL0437328,BL0437329, BL0437327, BL0437326, BL0437325 |
| Therapeutic Target Database | T89988 |
| ChEMBL | CHEMBL4015 |
| IUPHAR | 59 |
| DrugBank | N/A |
| Name | TAK-779 |
|---|---|
| Molecular formula | C33H39ClN2O2 |
| IUPAC name | dimethyl-[[4-[[3-(4-methylphenyl)-8,9-dihydro-7H-benzo[7]annulene-6-carbonyl]amino]phenyl]methyl]-(oxan-4-yl)azanium;chloride |
| Molecular weight | 531.137 |
| Hydrogen bond acceptor | 3 |
| Hydrogen bond donor | 1 |
| XlogP | None |
| Synonyms | HY-13406 TAK 779 BQW1Y9KIIP N,N-dimethyl-N-(4-(((2-(4-methylphenyl)-6,7-dihydro-5H-benzocyclohepten-8-yl)carbonyl)amino)benzyl)-N-(4-tetrahydropyranyl)-ammonium chloride UNII-BQW1Y9KIIP [ Show all ] |
| Inchi Key | VDALIBWXVQVFGZ-UHFFFAOYSA-N |
| Inchi ID | InChI=1S/C33H38N2O2.ClH/c1-24-7-11-27(12-8-24)28-14-13-26-5-4-6-29(22-30(26)21-28)33(36)34-31-15-9-25(10-16-31)23-35(2,3)32-17-19-37-20-18-32;/h7-16,21-22,32H,4-6,17-20,23H2,1-3H3;1H |
| PubChem CID | 183789 |
| ChEMBL | CHEMBL41275 |
| IUPHAR | N/A |
| BindingDB | N/A |
| DrugBank | N/A |
Structure | 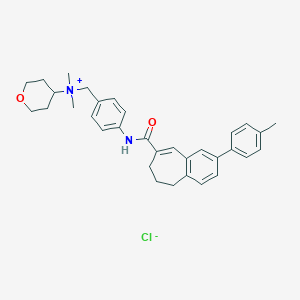 |
| Lipinski's druglikeness | This ligand is heavier than 500 daltons. Partition coefficient log P of this ligand is not available. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| IC50 | 0.03 nM | PMID17715128 | ChEMBL |
| IC50 | 0.08 nM | PMID17715128 | ChEMBL |
| IC50 | 0.25 nM | PMID17715128 | ChEMBL |
| IC50 | 0.95 nM | PMID25766632 | ChEMBL |
| IC50 | 27.0 nM | PMID10969972, PMID10821717, Med Chem Res, (2011) 20:9:1704, PMID22931505, PMID12954060, PMID11931608 | ChEMBL |
| IC50 | 28.0 nM | PMID19237282 | ChEMBL |
| Inhibition | 38.1 % | PMID17715128 | ChEMBL |
| Inhibition | 90.7 % | PMID17715128 | ChEMBL |
| Inhibition | 91.3 % | PMID17715128 | ChEMBL |
| Ki | 2.0 nM | PMID25766632 | ChEMBL |
| Ki | 2.63 nM | PMID12954060 | ChEMBL |
| Ki | 4.169 nM | PMID12954060 | ChEMBL |
| Ki | 6.918 nM | PMID12954060 | ChEMBL |
| Ki | 9.772 nM | PMID12954060 | ChEMBL |
| Ki | 12.59 nM | PMID12954060 | ChEMBL |
| Ki | 17.78 nM | PMID12954060 | ChEMBL |
| Ki | 21.38 nM | PMID12954060 | ChEMBL |
| Ki | 27.54 nM | PMID12954060 | ChEMBL |
| Ki | 28.84 nM | PMID12954060 | ChEMBL |
| Ki | 104.71 nM | PMID12954060 | ChEMBL |
| Ki | 147.91 nM | PMID12954060 | ChEMBL |
| Ki | 181.97 nM | PMID12954060 | ChEMBL |
| Ki | 1348.96 nM | PMID12954060 | ChEMBL |
| pKb | 9.53 - | PMID12954060 | ChEMBL |
zhanglab![]() zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218