

You can:
| Name | 316173-57-6 |
|---|---|
| Molecular formula | C20H19N7O2 |
| IUPAC name | 4-(furan-2-yl)-10-[3-(4-methoxyphenyl)propyl]-3,5,6,8,10,11-hexazatricyclo[7.3.0.02,6]dodeca-1(9),2,4,7,11-pentaen-7-amine |
| Molecular weight | 389.419 |
| Hydrogen bond acceptor | 7 |
| Hydrogen bond donor | 1 |
| XlogP | 2.7 |
| Synonyms | AX8233433 CTK8B9468 MFCD08703126 SCH442,416 2-(2-Furanyl)-7-[3-(4-methoxyphenyl)propyl]-7H-pyrazolo [4,3-e][1,2,4]triazolo[1,5-c]pyrimidin-5-amine; 5-amino-7-(3-(4-methoxyphenyl)propyl)-2-(2 furyl)pyrazolo[4,3-e]-1,2,4-triazolo[1,5-c]pyrimidine [ Show all ] |
| Inchi Key | AEULVFLPCJOBCE-UHFFFAOYSA-N |
| Inchi ID | InChI=1S/C20H19N7O2/c1-28-14-8-6-13(7-9-14)4-2-10-26-18-15(12-22-26)19-23-17(16-5-3-11-29-16)25-27(19)20(21)24-18/h3,5-9,11-12H,2,4,10H2,1H3,(H2,21,24) |
| PubChem CID | 10668061 |
| ChEMBL | CHEMBL136689 |
| IUPHAR | 3283 |
| BindingDB | 50094037 |
| DrugBank | N/A |
Structure | 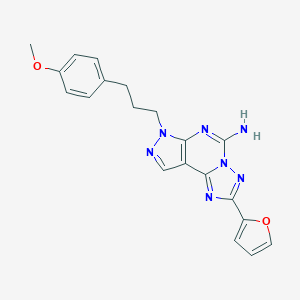 |
| Lipinski's druglikeness | This ligand satisfies Lipinski's rule of five. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| INH | 0.048 nM | PMID21214204 | ChEMBL |
| Kd | 1.956 nM | DOI: http://dx.doi.org/10.6019/CHEMBL3885741 | ChEMBL |
| Kd | 4.66 nM | DOI: http://dx.doi.org/10.6019/CHEMBL3885741 | ChEMBL |
| Ki | <0.048 nM | PMID20937560 | ChEMBL |
| Ki | <0.048 nM | PMID20937560 | BindingDB |
| Ki | 0.048 nM | PMID11087559, PMID24972108 | BindingDB,ChEMBL |
| Ki | 0.048 nM | PMID24972108 | BindingDB |
| Ki | 0.0501187 - 3.98107 nM | PMID11087559, PMID20801028 | IUPHAR |
| Ki | 4.1 nM | PMID23200243, PMID20801028, PMID21185184 | BindingDB,ChEMBL |
| Ki | 11.1 nM | PMID23200243 | ChEMBL |
| koff | 0.4149 min^-1 | DOI: http://dx.doi.org/10.6019/CHEMBL3885741 | ChEMBL |
| kon | 0.222 nM^-1 min^-1 | DOI: http://dx.doi.org/10.6019/CHEMBL3885741 | ChEMBL |
| kon | 726200.0 Ms-1 | DOI: http://dx.doi.org/10.6019/CHEMBL3885741 | ChEMBL |
| kon | 793267.0 Ms-1 | DOI: http://dx.doi.org/10.6019/CHEMBL3885741 | ChEMBL |
| k_off | 0.00132 s-1 | DOI: http://dx.doi.org/10.6019/CHEMBL3885741 | ChEMBL |
| k_off | 0.003348 s-1 | DOI: http://dx.doi.org/10.6019/CHEMBL3885741 | ChEMBL |
zhanglab![]() zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218