| Synonyms | IDI1_000511
Anatensol (Salt/Mix)
KBio3_002320
MRF-0000359
BIDD:PXR0184
Lopac0_000555
NCGC00015436-08
BSPBio_003100
Moditen (Tabl or elixir)
Omca (TN)
PLDUPXSUYLZYBN-UHFFFAOYSA-N
D0B7SP
1-Piperazineethanol, 4-(3-(2-(trifluoromethyl)phenothiazin-10-yl)propyl)-
Prolixin (Salt/Mix)
Decanoate (TN)
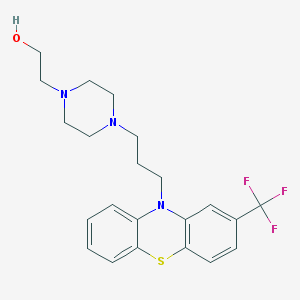
2-(4-(3-(2-(trifluoromethyl)-10H-phenothiazin-10-yl)propyl)piperazin-1-yl)ethanol
SBI-0050538.P003
Elinol
2-[4-[3-[2-(trifluoromethyl)-10-phenothiazinyl]propyl]-1-piperazinyl]ethanol;hydrochloride
Siqualon
Fluorfenazine
4-(3-(2-(trifluoromethyl)phenothiazin-10-yl)propyl)-1-Piperazineethanol
SQ 4918
Fluphenazinum
A836405
UNII-S79426A41Z
ZINC19203912
KBio2_002386
Apo-Fluphenazine
KBioGR_002386
NCGC00015436-03
BPBio1_001187
MCULE-7579992672
NCGC00024429-03
Calmansial
Moditen Hcl
Permitil (Salt/Mix)
cid_3372
1-(2-Hydroxyethyl)-4-(3-(trifluoromethyl-10-phenothiazinyl)propyl)-piperazine
Prestwick2_000320
Dapotum (TN)
10-(3'-(4"-(beta-Hydroxyethyl)-1"-piperazinyl)-propyl)-3-trifluoromethylphenothiazine
Prolixine
DSSTox_CID_3068
2-(4-{3-[2-(trifluoromethyl)-10H-phenothiazin-10-yl]propyl}piperazin-1-yl)ethan-1-ol
Selecten (TN)
Flufenazin
2-[4-[3-[2-(trifluoromethyl)phenothiazin-10-yl]propyl]piperazin-1-yl]ethanol;hydrochloride
Spectrum2_001199
Fluorphenazine
4-[3-[2-(Trifluoromethyl)-10H-phenothiazin-10-yl]propyl]-1-piperazineethanol
Tensofin (Salt/Mix)
GTPL204
AB00053587_19
AKOS001659031
KBio2_005905
BDBM78433
L000257
NCGC00015436-06
BRD-K55127134-300-07-0
Modecate (TN)
NSC 62323
CCG-204645
Phenothiazine, 10-(3-(4-(2-hydroxyethyl)-1-piperazinyl)propyl)-2-(trifluoromethyl)-
CTK8F9822
1-Piperazineethanol, 4-(3-(2-(trifluoromethyl)-10H-phenothiazin-10-yl)propyl)- (9CI)
Probes2_000295
1451-EP2272537A2
S79426A41Z
DTXSID2023068
2-(Trifluoromethyl)-10-(3-(1-(beta-hydroxyethyl)-4-piperazinyl)propyl)phenothiazine
Sinqualone (TN)
flufenazine
4-(3-(-Trifluoromethyl-10-phenothiazyl)-propyl)-1-piperazineethanol
Spectrum5_001536
Fluphenazine (INN)
Trancin (TN)
HSDB 3334
KBio1_000511
Anatensol (TN)
KBio3_002865
NCGC00015436-01
Biomol-NT_000023
LS-112565
NCGC00015436-09
C07010
Moditen (TN)
Pacinol
CHEMBL726
Prestwick0_000320
D0P5SA
1-Piperazineethanol, 4-[3-[2-(trifluoromethyl)-10H-phenothiazin-10-yl]propyl]-
Prolixin (TN)
Deconoate (TN)
2-(4-(3-[2-(Trifluoromethyl)-10H-phenothiazin-10-yl]propyl)-1-piperazinyl)ethanol
SCHEMBL19221
Enanthate (TN)
2-[4-[3-[2-(trifluoromethyl)phenothiazin-10-yl]propyl]piperazin-1-yl]ethanol
SPBio_001277
Fluorfenazine pound (3/4)<4-(3-(2-Trifluoromethyl-10-phenothiazyl)-propyl)-1-piperazineethanol
SR-01000003048-5
Fluphenazinum [INN-Latin]
AB00053587
Vespazine
AC1L1FSB
KBio2_003337
AX8130619
KBioSS_000769
NCGC00015436-04
BRD-K55127134-001-01-7
MFCD00242759
NCGC00024429-04
CAS-146-56-5
MolPort-001-727-938
Permitil (TN)
cid_6602611
1-(2-Hydroxyethyl)-4-[3-(trifluoromethyl-10-phenothiazinyl)propyl]piperazine
Prestwick3_000320
Dapotum D (TN)
10-(3'-(4''-(beta-Hydroxyethyl)-1''-piperazinyl)-propyl)-3-trifluoromethylphenothiazine
Prolixine (Salt/Mix)
DSSTox_GSID_23068
2-(4-{3-[2-(trifluoromethyl)-10H-phenothiazin-10-yl]propyl}piperazin-1-yl)ethanol
Sevinol
Flufenazina
2-[4-[3-[2-(trifluoromethyl)phenothiazin-10-yl]propyl]piperazino]ethanol;hydrochloride
Spectrum3_001420
Fluphenazin
47646-09-3
Tox21_110146
HMS2089C16
Anatensol
KBio2_007522
BG0198
Lopac-F-4765
NCGC00015436-07
BSPBio_000320
Moditen
NSC62323
CHEBI:5123
Phthorphenazine
D07977
1-Piperazineethanol, 4-(3-(2-(trifluoromethyl)-10H-phenothiazin-10-yl)propyl)-(9CI)
Prolixin
DB00623
1451-EP2298776A1
S94
EINECS 200-702-9
2-[4-[3-[2-(trifluoromethyl)-10-phenothiazinyl]propyl]-1-piperazinyl]ethanol
Siqualine
Flunanthate (TN)
4-(3-(2-(Trifluoromethyl)-10H-phenothiazin-10-yl)propyl)-1-piperazineethanol
Spectrum_000289
Fluphenazine [INN:BAN]
A 4077
Triflumethazine
Hydrochloride, Fluphenazine
Yespazine
KBio2_000769
API0006719
KBioGR_001375
NCGC00015436-02
BPBio1_000352
Lyogen (TN)
NCGC00015436-11
C22H26F3N3OS
Moditen Enanthate Injection (TN)
Permitil
ChemDiv1_024180
0680AC
Prestwick1_000320
Dapotum
1-Piperazineethanol, 4-[3-[2-(trifluoromethyl)phenothiazin-10-yl]propyl]-
Prolixin Concentrate
DivK1c_000511
2-(4-(3-[2-(Trifluoromethyl)-10H-phenothiazin-10-yl]propyl)-1-piperazinyl)ethanol #
Sediten (TN)
Fludecate (TN)
2-[4-[3-[2-(trifluoromethyl)phenothiazin-10-yl]propyl]piperazin-1-yl]ethanol dihydrochloride
SPBio_002539
Fluorophenazine
4-[3-(2-Trifluoromethyl-10-phenothiazyl)-propyl]-1-piperazineethanol
STK050763
Ftorphenazine
AB00053587-18
AJ-72569
KBio2_004954
BCP21399
KBioSS_002391
NCGC00015436-05
BRD-K55127134-300-05-4
MLS001076508
NINDS_000511
CAS-69-23-8
Permitil Concentrate
cMAP_000035
1-Piperazineethanol, 4-(3-(2-(trifluoromethyl)-10H-phenothiazin-10-yl)propyl)-
Probes1_000254
Dapotum Injektion (TN)
10-(3-(2-Hydroxyethyl)piperazinopropyl)-2-(trifluoromethyl)phenothiazine
S 94
DSSTox_RID_76862
2-(Trifluoromethyl)-10-(3-(1-(beta-hydroxyethyl)-4-pipe razinyl)propyl)phenothiazine
Sevinol (TN)
Flufenazina [DCIT]
Spectrum4_000828
69-23-8
Tox21_110146_1
HMS655L02 [ Show all ] |
|---|


![]() zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218