

You can:
| Name | G-protein coupled receptor 35 |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | GPR35 |
| Synonym | G-protein coupled receptor 3 GPR35 KYNA receptor Kynurenic acid receptor |
| Disease | N/A |
| Length | 309 |
| Amino acid sequence | MNGTYNTCGSSDLTWPPAIKLGFYAYLGVLLVLGLLLNSLALWVFCCRMQQWTETRIYMTNLAVADLCLLCTLPFVLHSLRDTSDTPLCQLSQGIYLTNRYMSISLVTAIAVDRYVAVRHPLRARGLRSPRQAAAVCAVLWVLVIGSLVARWLLGIQEGGFCFRSTRHNFNSMAFPLLGFYLPLAVVVFCSLKVVTALAQRPPTDVGQAEATRKAARMVWANLLVFVVCFLPLHVGLTVRLAVGWNACALLETIRRALYITSKLSDANCCLDAICYYYMAKEFQEASALAVAPSAKAHKSQDSLCVTLA |
| UniProt | Q9HC97 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | Q9HC97 |
| 3D structure model | This predicted structure model is from GPCR-EXP Q9HC97. |
| BioLiP | N/A |
| Therapeutic Target Database | N/A |
| ChEMBL | CHEMBL1293267 |
| IUPHAR | 102 |
| DrugBank | BE0005562 |
| Name | MLS000575039 |
|---|---|
| Molecular formula | C24H22N2O5S2 |
| IUPAC name | 2-hydroxy-4-[4-[(5Z)-5-[(E)-2-methyl-3-phenylprop-2-enylidene]-4-oxo-2-sulfanylidene-1,3-thiazolidin-3-yl]butanoylamino]benzoic acid |
| Molecular weight | 482.569 |
| Hydrogen bond acceptor | 7 |
| Hydrogen bond donor | 3 |
| XlogP | 5.1 |
| Synonyms | 2-hydroxy-4-[4-[(5Z)-4-keto-5-[(E)-2-methyl-3-phenyl-prop-2-enylidene]-2-thioxo-thiazolidin-3-yl]butanoylamino]benzoic acid MolPort-000-506-071 4-[4-[(5Z)-5-[(E)-2-methyl-3-phenyl-prop-2-enylidene]-4-oxidanylidene-2-sulfanylidene-1,3-thiazolidin-3-yl]butanoylamino]-2-oxidanyl-benzoic acid ML 145 STL227169 [ Show all ] |
| Inchi Key | COFMYJWNXSFLKQ-QIROLCGISA-N |
| Inchi ID | InChI=1S/C24H22N2O5S2/c1-15(12-16-6-3-2-4-7-16)13-20-22(29)26(24(32)33-20)11-5-8-21(28)25-17-9-10-18(23(30)31)19(27)14-17/h2-4,6-7,9-10,12-14,27H,5,8,11H2,1H3,(H,25,28)(H,30,31)/b15-12+,20-13- |
| PubChem CID | 2286812 |
| ChEMBL | CHEMBL1384502 |
| IUPHAR | N/A |
| BindingDB | 50185 |
| DrugBank | N/A |
Structure | 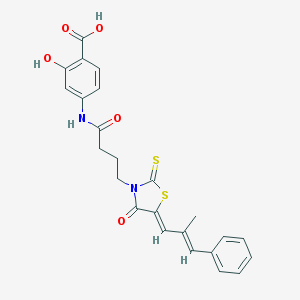 |
| Lipinski's druglikeness | This ligand has a partition coefficient log P greater than 5. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| EC50 | 522.0 nM | PMID23888932 | BindingDB |
| IC50 | 27.0 nM | PMID23888932 | BindingDB,ChEMBL |
| IC50 | 522.0 nM | PMID23888932 | ChEMBL |
| IC50 | 2361.0 nM | , PubChem BioAssay data set | BindingDB,ChEMBL |
| Ki | 8.76 nM | PMID23888932 | ChEMBL |
| Ki | 8.8 nM | PMID23888932 | BindingDB |
zhanglab![]() zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218