

You can:
| Name | Alpha-2A adrenergic receptor |
|---|---|
| Species | Rattus norvegicus (Rat) |
| Gene | Adra2a |
| Synonym | CA2-47 alpha2D alpha2A-AR alpha2A-adrenoceptor alpha2A-adrenergic receptor [ Show all ] |
| Disease | N/A for non-human GPCRs |
| Length | 450 |
| Amino acid sequence | MGSLQPDAGNSSWNGTEAPGGGTRATPYSLQVTLTLVCLAGLLMLFTVFGNVLVIIAVFTSRALKAPQNLFLVSLASADILVATLVIPFSLANEVMGYWYFGKVWCEIYLALDVLFCTSSIVHLCAISLDRYWSITQAIEYNLKRTPRRIKAIIVTVWVISAVISFPPLISIEKKGAGGGQQPAEPSCKINDQKWYVISSSIGSFFAPCLIMILVYVRIYQIAKRRTRVPPSRRGPDACSAPPGGADRRPNGLGPERGAGTAGAEAEPLPTQLNGAPGEPAPTRPRDGDALDLEESSSSEHAERPQGPGKPERGPRAKGKTKASQVKPGDSLPRRGPGAAGPGASGSGQGEERAGGAKASRWRGRQNREKRFTFVLAVVIGVFVVCWFPFFFTYTLIAVGCPVPYQLFNFFFWFGYCNSSLNPVIYTIFNHDFRRAFKKILCRGDRKRIV |
| UniProt | P22909 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | N/A |
| 3D structure model | No available structures or models |
| BioLiP | N/A |
| Therapeutic Target Database | N/A |
| ChEMBL | CHEMBL327 |
| IUPHAR | N/A |
| DrugBank | N/A |
| Name | DEXMEDETOMIDINE |
|---|---|
| Molecular formula | C13H16N2 |
| IUPAC name | 5-[(1S)-1-(2,3-dimethylphenyl)ethyl]-1H-imidazole |
| Molecular weight | 200.285 |
| Hydrogen bond acceptor | 1 |
| Hydrogen bond donor | 1 |
| XlogP | 3.1 |
| Synonyms | CHEBI:4466 DB00633 Dexmedetomidine [USAN:INN:BAN] MolPort-035-395-718 SC-78886 [ Show all ] |
| Inchi Key | CUHVIMMYOGQXCV-NSHDSACASA-N |
| Inchi ID | InChI=1S/C13H16N2/c1-9-5-4-6-12(10(9)2)11(3)13-7-14-8-15-13/h4-8,11H,1-3H3,(H,14,15)/t11-/m0/s1 |
| PubChem CID | 5311068 |
| ChEMBL | CHEMBL778 |
| IUPHAR | 521 |
| BindingDB | 50085683 |
| DrugBank | DB00633 |
Structure | 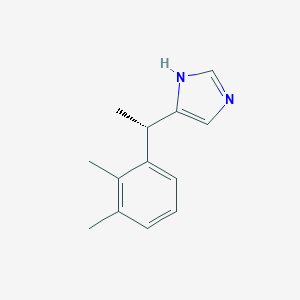 |
| Lipinski's druglikeness | This ligand satisfies Lipinski's rule of five. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| Ki | 0.015 nM | PMID10715142 | BindingDB |
| Ki | 0.015 nM | PMID10715142, PMID10753480 | BindingDB,ChEMBL |
| Ki | 5.0 nM | PMID10753480 | BindingDB,ChEMBL |
zhanglab![]() zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218