

You can:
| Name | 5-hydroxytryptamine receptor 1A |
|---|---|
| Species | Rattus norvegicus (Rat) |
| Gene | Htr1a |
| Synonym | 5-HT1A receptor 5-hydroxytryptamine (serotonin) receptor 1A, G protein-coupled 5-HT1A ADRB2RL1 ADRBRL1 [ Show all ] |
| Disease | N/A for non-human GPCRs |
| Length | 422 |
| Amino acid sequence | MDVFSFGQGNNTTASQEPFGTGGNVTSISDVTFSYQVITSLLLGTLIFCAVLGNACVVAAIALERSLQNVANYLIGSLAVTDLMVSVLVLPMAALYQVLNKWTLGQVTCDLFIALDVLCCTSSILHLCAIALDRYWAITDPIDYVNKRTPRRAAALISLTWLIGFLISIPPMLGWRTPEDRSDPDACTISKDHGYTIYSTFGAFYIPLLLMLVLYGRIFRAARFRIRKTVRKVEKKGAGTSLGTSSAPPPKKSLNGQPGSGDWRRCAENRAVGTPCTNGAVRQGDDEATLEVIEVHRVGNSKEHLPLPSESGSNSYAPACLERKNERNAEAKRKMALARERKTVKTLGIIMGTFILCWLPFFIVALVLPFCESSCHMPALLGAIINWLGYSNSLLNPVIYAYFNKDFQNAFKKIIKCKFCRR |
| UniProt | P19327 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | N/A |
| 3D structure model | No available structures or models |
| BioLiP | N/A |
| Therapeutic Target Database | N/A |
| ChEMBL | CHEMBL273 |
| IUPHAR | 1 |
| DrugBank | N/A |
| Name | clozapine |
|---|---|
| Molecular formula | C18H19ClN4 |
| IUPAC name | 3-chloro-6-(4-methylpiperazin-1-yl)-11H-benzo[b][1,4]benzodiazepine |
| Molecular weight | 326.828 |
| Hydrogen bond acceptor | 3 |
| Hydrogen bond donor | 1 |
| XlogP | 3.1 |
| Synonyms | AN-16004 GTPL38 NCGC00022902-09 BDBM22869 HMS1921C16 [ Show all ] |
| Inchi Key | QZUDBNBUXVUHMW-UHFFFAOYSA-N |
| Inchi ID | InChI=1S/C18H19ClN4/c1-22-8-10-23(11-9-22)18-14-4-2-3-5-15(14)20-16-7-6-13(19)12-17(16)21-18/h2-7,12,20H,8-11H2,1H3 |
| PubChem CID | 135398737 |
| ChEMBL | CHEMBL42 |
| IUPHAR | 38 |
| BindingDB | 50001884 |
| DrugBank | DB00363 |
Structure | 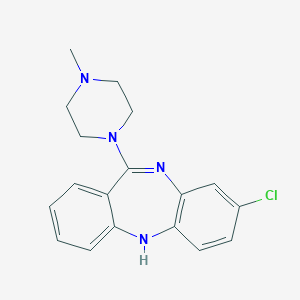 |
| Lipinski's druglikeness | This ligand satisfies Lipinski's rule of five. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| IC50 | 267.6 nM | PMID12361392 | BindingDB,ChEMBL |
| IC50 | 491.0 nM | DrugMatrix in vitro pharmacology data | ChEMBL |
| IC50 | 580.0 nM | PMID1672156 | BindingDB,ChEMBL |
| IC50 | 600.0 nM | PMID1676427 | BindingDB,ChEMBL |
| IC50 | 1010.0 nM | PMID8831770 | BindingDB,ChEMBL |
| IC50 | 1015.5 nM | PMID7914536 | BindingDB,ChEMBL |
| IC50 | 2000.0 nM | PMID7914539, PMID8568802, PMID8676355 | BindingDB,ChEMBL |
| Ki | 38.5 nM | PMID7473548 | ChEMBL |
| Ki | 39.0 nM | PMID1346653, PMID7473548, PMID7473547 | BindingDB,ChEMBL |
| Ki | 95.5 nM | PMID17870534 | ChEMBL |
| Ki | 111.0 nM | PMID9622541, PMID7909336 | BindingDB,ChEMBL |
| Ki | 130.0 nM | PMID8709107 | BindingDB,ChEMBL |
| Ki | 140.0 nM | PMID21816515 | BindingDB,ChEMBL |
| Ki | 141.6 nM | PMID23675993 | ChEMBL |
| Ki | 150.0 nM | PMID8691438 | BindingDB,ChEMBL |
| Ki | 160.0 nM | PMID19072656, PMID20481570 | BindingDB,ChEMBL |
| Ki | 161.0 nM | PMID26483200 | BindingDB,ChEMBL |
| Ki | 185.6 nM | PMID24487191, MedChemComm, (2015) 6:5:831 | ChEMBL |
| Ki | 247.0 nM | PMID9876110 | ChEMBL |
| Ki | 247.0 nM | PMID9876110 | BindingDB |
| Ki | 260.0 nM | PMID23353740, PMID23792350 | BindingDB,ChEMBL |
| Ki | 281.0 nM | DrugMatrix in vitro pharmacology data | ChEMBL |
| Ki | 380.0 nM | PMID20153652 | BindingDB,ChEMBL |
| Ki | 415.0 nM | PMID10464021 | BindingDB |
| Ki | 415.0 nM | PMID10464021 | ChEMBL |
| Ki | 443.0 nM | PMID10649982 | BindingDB |
| Ki | 443.0 nM | PMID10649982 | ChEMBL |
| Ki | 640.0 nM | PMID1353116 | BindingDB,ChEMBL |
zhanglab![]() zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218