

You can:
| Name | Mu-type opioid receptor |
|---|---|
| Species | Cavia porcellus (Guinea pig) |
| Gene | OPRM1 |
| Synonym | M-OR-1 MOR-1 |
| Disease | N/A for non-human GPCRs |
| Length | 98 |
| Amino acid sequence | YTKMKTATNIYIFNLALADALATSTLPFQSVNYLMGTWPFGTILCKIVISIDYYNMFTSIFTLCTMSVDRYIAVCHPVKALDFRTPRNAKTVNVCNWI |
| UniProt | P97266 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | N/A |
| 3D structure model | No available structures or models |
| BioLiP | N/A |
| Therapeutic Target Database | N/A |
| ChEMBL | CHEMBL4354 |
| IUPHAR | N/A |
| DrugBank | N/A |
| Name | naltrexone |
|---|---|
| Molecular formula | C20H23NO4 |
| IUPAC name | (4R,4aS,7aR,12bS)-3-(cyclopropylmethyl)-4a,9-dihydroxy-2,4,5,6,7a,13-hexahydro-1H-4,12-methanobenzofuro[3,2-e]isoquinolin-7-one |
| Molecular weight | 341.407 |
| Hydrogen bond acceptor | 5 |
| Hydrogen bond donor | 2 |
| XlogP | 1.9 |
| Synonyms | Vivitrol DQCKKXVULJGBQN-XFWGSAIBSA-N GTPL1639 MCULE-7032963403 Naltrel [ Show all ] |
| Inchi Key | DQCKKXVULJGBQN-XFWGSAIBSA-N |
| Inchi ID | InChI=1S/C20H23NO4/c22-13-4-3-12-9-15-20(24)6-5-14(23)18-19(20,16(12)17(13)25-18)7-8-21(15)10-11-1-2-11/h3-4,11,15,18,22,24H,1-2,5-10H2/t15-,18+,19+,20-/m1/s1 |
| PubChem CID | 5360515 |
| ChEMBL | CHEMBL19019 |
| IUPHAR | 1639 |
| BindingDB | 60212, 50000787 |
| DrugBank | DB00704 |
Structure | 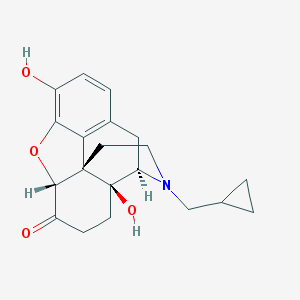 |
| Lipinski's druglikeness | This ligand satisfies Lipinski's rule of five. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| Dose ratio | 276.0 - | PMID2409281 | ChEMBL |
| IC50 | 0.2 nM | PMID7853350 | ChEMBL |
| IC50 | 0.2 nM | PMID7853350 | BindingDB |
| IC50 | 0.23 nM | PMID7996538 | ChEMBL |
| IC50 | 0.23 nM | PMID7996538 | BindingDB |
| IC50 | 0.73 nM | PMID1335078, PMID1320121 | BindingDB |
| IC50 | 0.73 nM | PMID1335078, PMID1320121 | ChEMBL |
| IC50 ratio | 98.0 - | PMID2160538 | ChEMBL |
| Ke | 0.2 nM | PMID14736254 | ChEMBL |
| Ke | 0.93 nM | PMID12723940 | ChEMBL |
| Ke | 1.0 - | PMID1851846 | ChEMBL |
| Ke | 1.0 nM | PMID2160538, PMID2832604, PMID1648136, PMID2838632 | ChEMBL |
| Ke | 1.2 nM | PMID8627605 | ChEMBL |
| Ki | 0.17 nM | PMID12749896, PMID11425545 | ChEMBL |
| Ki | 0.17 nM | PMID12749896, PMID11425545 | BindingDB |
| Ki | 0.325 nM | PMID19349178 | ChEMBL |
| Ki | 0.335 nM | PMID20483601, PMID20056539, PMID21641219, PMID18755589, PMID19962305 | ChEMBL |
| Ki | 0.37 nM | PMID2879914 | ChEMBL |
| Ki | 0.37 nM | PMID2879914 | BindingDB |
| Ki | 0.4 nM | PMID17004724, PMID19253983 | BindingDB |
| Ki | 0.4 nM | PMID17004724, PMID19253983, PMID9686407 | BindingDB,ChEMBL |
| Ki | 0.56 nM | PMID9873693 | ChEMBL |
| Ki | 0.56 nM | PMID9873693 | BindingDB |
| Ki | 0.75 nM | PMID8627605 | BindingDB |
| Ki | 0.75 nM | PMID8627605 | ChEMBL |
| Ki | 0.8 nM | PMID2160538, PMID1648136 | BindingDB |
| Ki | 0.8 nM | PMID9057861, PMID2160538, PMID1648136 | ChEMBL |
| Ki | 0.8 nM | PMID9057861 | BindingDB |
| Ki | 0.93 nM | PMID9599247, PMID9873693, PMID9857089 | BindingDB |
| Ki | 0.93 nM | PMID9767649, PMID9873693, PMID9599247, PMID9857089 | BindingDB,ChEMBL |
| Ki | 1.08 nM | PMID2409281 | ChEMBL |
| Ki | 1.1 nM | PMID2409281 | BindingDB |
| Ki | 1.39 nM | PMID12723940, PMID9857089 | ChEMBL |
| Ki | 1.4 nM | PMID12723940, PMID9857089 | BindingDB |
| Ki | 7.6 - | PMID1851846 | ChEMBL |
| Ratio | 0.02 - | PMID1851846 | ChEMBL |
zhanglab![]() zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218