

You can:
| Name | Melatonin receptor type 1B |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | MTNR1B |
| Synonym | MT2 receptor mel1b receptor Mel-1B-R |
| Disease | Epilepsy |
| Length | 362 |
| Amino acid sequence | MSENGSFANCCEAGGWAVRPGWSGAGSARPSRTPRPPWVAPALSAVLIVTTAVDVVGNLLVILSVLRNRKLRNAGNLFLVSLALADLVVAFYPYPLILVAIFYDGWALGEEHCKASAFVMGLSVIGSVFNITAIAINRYCYICHSMAYHRIYRRWHTPLHICLIWLLTVVALLPNFFVGSLEYDPRIYSCTFIQTASTQYTAAVVVIHFLLPIAVVSFCYLRIWVLVLQARRKAKPESRLCLKPSDLRSFLTMFVVFVIFAICWAPLNCIGLAVAINPQEMAPQIPEGLFVTSYLLAYFNSCLNAIVYGLLNQNFRREYKRILLALWNPRHCIQDASKGSHAEGLQSPAPPIIGVQHQADAL |
| UniProt | P49286 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | P49286 |
| 3D structure model | This predicted structure model is from GPCR-EXP P49286. |
| BioLiP | N/A |
| Therapeutic Target Database | T48268 |
| ChEMBL | CHEMBL1946 |
| IUPHAR | 288 |
| DrugBank | BE0000327 |
| Name | Melatonin |
|---|---|
| Molecular formula | C13H16N2O2 |
| IUPAC name | N-[2-(5-methoxy-1H-indol-3-yl)ethyl]acetamide |
| Molecular weight | 232.283 |
| Hydrogen bond acceptor | 2 |
| Hydrogen bond donor | 2 |
| XlogP | 0.8 |
| Synonyms | NCGC00015680-10 Tox21_500787 NCGC00090727-01 [3H]MLT NCGC00090727-09 [ Show all ] |
| Inchi Key | DRLFMBDRBRZALE-UHFFFAOYSA-N |
| Inchi ID | InChI=1S/C13H16N2O2/c1-9(16)14-6-5-10-8-15-13-4-3-11(17-2)7-12(10)13/h3-4,7-8,15H,5-6H2,1-2H3,(H,14,16) |
| PubChem CID | 896 |
| ChEMBL | CHEMBL45 |
| IUPHAR | 1357, 224 |
| BindingDB | 9019 |
| DrugBank | DB01065 |
Structure | 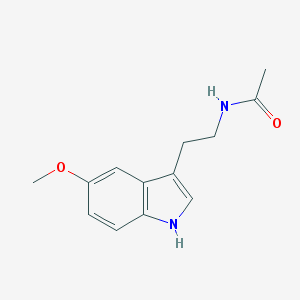 |
| Lipinski's druglikeness | This ligand satisfies Lipinski's rule of five. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| N/A | N/A | DrugBank | |
| EC50 | 0.069 nM | PMID26023814 | ChEMBL |
| EC50 | 0.069 nM | PMID26023814 | BindingDB |
| EC50 | 0.42 nM | PMID21420861 | BindingDB,ChEMBL |
| EC50 | 0.47 nM | PMID12061881 | BindingDB,ChEMBL |
| EC50 | 0.49 nM | PMID18778943, PMID12646022, PMID20444610 | BindingDB,ChEMBL |
| EC50 | 0.82 nM | PMID21568291 | BindingDB,ChEMBL |
| EC50 | 0.8511 nM | PMID21473625 | ChEMBL |
| ECmax | 102.0 % | PMID21420861 | ChEMBL |
| Emax | 100.0 % | PMID12061881 | ChEMBL |
| Emax | 104.0 % | PMID12646022, PMID20444610 | ChEMBL |
| IA | 1.0 - | PMID15943478 | ChEMBL |
| IAr | 1.0 - | PMID15293992, PMID11520198 | ChEMBL |
| IC50 | 0.3 nM | PMID12643943 | BindingDB |
| IC50 | 0.3 nM | PMID12643943 | ChEMBL |
| IC50 | 0.53 nM | PMID21377769 | BindingDB |
| IC50 | 0.53 nM | PMID21377769, PMID18372181 | BindingDB,ChEMBL |
| Inhibition | 100.0 % | PMID26367450 | ChEMBL |
| Intrinsic activity | 1.0 - | PMID12672242 | ChEMBL |
| Intrinsic activity | 1.01 - | PMID18052314 | ChEMBL |
| Kd | 0.15 nM | MedChemComm, (2015) 6:7:1340 | ChEMBL |
| Kd | 0.28 - 0.912 nM | PMID10696085 | IUPHAR |
| Ki | 0.12 nM | PMID9435890 | BindingDB,ChEMBL |
| Ki | 0.15 nM | PMID26023814, PMID26367450 | BindingDB |
| Ki | 0.15 nM | PMID26023814, PMID26367450 | ChEMBL |
| Ki | 0.158489 - 0.398108 nM | PMID12764576, PMID9089668, PMID2991499 | IUPHAR |
| Ki | 0.18 nM | PMID9089668 | PDSP,BindingDB |
| Ki | 0.19 nM | PMID9618428 | PDSP,BindingDB |
| Ki | 0.195 nM | PMID12213063 | BindingDB,ChEMBL |
| Ki | 0.21 nM | PMID21473625, PMID21568291 | BindingDB,ChEMBL |
| Ki | 0.23 nM | PMID7946354, Bioorg. Med. Chem. Lett., (1997) 7:17:2177 | BindingDB,ChEMBL |
| Ki | 0.23 nM | N/A | BindingDB |
| Ki | 0.2399 nM | PMID18052314 | ChEMBL |
| Ki | 0.25 nM | PMID10455277 | PDSP,BindingDB |
| Ki | 0.257 nM | PMID17149869 | ChEMBL |
| Ki | 0.2818 nM | PMID15293992, PMID11520198, PMID15943478 | ChEMBL |
| Ki | 0.29 nM | Bioorg. Med. Chem. Lett., (1997) 7:18:2409 | ChEMBL |
| Ki | 0.29 nM | N/A | BindingDB |
| Ki | 0.2951 nM | PMID12672242 | ChEMBL |
| Ki | 0.3 nM | PMID14643330, PMID14980664, PMID15203165 | BindingDB |
| Ki | 0.3 nM | PMID14643330, PMID14980664, PMID17481904, PMID15203165 | BindingDB,ChEMBL |
| Ki | 0.31 nM | PMID11960497, PMID12061881 | BindingDB,ChEMBL |
| Ki | 0.31 nM | PMID11960497 | BindingDB |
| Ki | 0.32 nM | PMID15013015 | ChEMBL |
| Ki | 0.32 nM | PMID15013015 | BindingDB |
| Ki | 0.33 nM | PMID9618903, PMID9840420, PMID10737738 | PDSP,BindingDB,ChEMBL |
| Ki | 0.3388 nM | PMID17459711, PMID21775151 | ChEMBL |
| Ki | 0.339 nM | PMID21775151 | BindingDB |
| Ki | 0.34 nM | PMID21764185, PMID26785296, PMID21420861 | BindingDB,ChEMBL |
| Ki | 0.34 nM | PMID26785296 | BindingDB |
| Ki | 0.35 nM | PMID16759094, PMID21392858 | BindingDB,ChEMBL |
| Ki | 0.3548 nM | PMID17346859 | ChEMBL |
| Ki | 0.38 nM | PMID9618428 | BindingDB |
| Ki | 0.41 nM | PMID18778943, PMID12646022, PMID25232966 | BindingDB,ChEMBL |
| Ki | 0.41 nM | PMID25232966 | BindingDB |
| Ki | 0.41 nM | PMID18778943 | PDSP |
| Ki | 0.429 nM | PMID23228808 | BindingDB |
| Ki | 0.429 nM | PMID20227878, PMID23228808 | BindingDB,ChEMBL |
| Ki | 0.48 nM | PMID9618903 | PDSP,BindingDB |
| Ki | 0.5 nM | PMID23265885 | ChEMBL |
| Ki | 0.5 nM | PMID23265885 | BindingDB |
| Ki | 0.52 nM | PMID26820449 | BindingDB |
| Ki | 0.52 nM | PMID26820449 | ChEMBL |
| Ki | 0.53 nM | PMID11063602 | BindingDB,ChEMBL |
| Ki | 0.6166 nM | PMID20674373 | ChEMBL |
| Ki | 0.617 nM | PMID20674373 | BindingDB |
| Ki | 0.7 nM | PMID15713384, PMID15380218 | BindingDB,ChEMBL |
| Ki | 0.741 nM | PMID18657980 | BindingDB,ChEMBL |
| Ki | 0.955 nM | MedChemComm, (2011) 2:10:991, MedChemComm, (2015) 6:7:1340, PMID19473848, PMID19193160 | ChEMBL |
| Ki | 1.11 nM | PMID7568007 | PDSP,BindingDB |
| pRA2 | 0.0 - | PMID12672242 | ChEMBL |
zhanglab![]() zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218