

You can:
| Name | Sphingosine 1-phosphate receptor 3 |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | S1PR3 |
| Synonym | Sphingosine 1-phosphate receptor Edg-3 S1P3 receptor S1P3 S1P receptor Edg-3 S1P receptor 3 [ Show all ] |
| Disease | Breast cancer |
| Length | 378 |
| Amino acid sequence | MATALPPRLQPVRGNETLREHYQYVGKLAGRLKEASEGSTLTTVLFLVICSFIVLENLMVLIAIWKNNKFHNRMYFFIGNLALCDLLAGIAYKVNILMSGKKTFSLSPTVWFLREGSMFVALGASTCSLLAIAIERHLTMIKMRPYDANKRHRVFLLIGMCWLIAFTLGALPILGWNCLHNLPDCSTILPLYSKKYIAFCISIFTAILVTIVILYARIYFLVKSSSRKVANHNNSERSMALLRTVVIVVSVFIACWSPLFILFLIDVACRVQACPILFKAQWFIVLAVLNSAMNPVIYTLASKEMRRAFFRLVCNCLVRGRGARASPIQPALDPSRSKSSSSNNSSHSPKVKEDLPHTAPSSCIMDKNAALQNGIFCN |
| UniProt | Q99500 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | Q99500 |
| 3D structure model | This predicted structure model is from GPCR-EXP Q99500. |
| BioLiP | N/A |
| Therapeutic Target Database | T11241 |
| ChEMBL | CHEMBL3892 |
| IUPHAR | 277 |
| DrugBank | N/A |
| Name | Sphingosine 1-phosphate |
|---|---|
| Molecular formula | C18H38NO5P |
| IUPAC name | [(E,2S,3R)-2-amino-3-hydroxyoctadec-4-enyl] dihydrogen phosphate |
| Molecular weight | 379.478 |
| Hydrogen bond acceptor | 6 |
| Hydrogen bond donor | 4 |
| XlogP | 1.9 |
| Synonyms | (2-amino-3-hydroxy-octadec-4-enoxy)phosphonic acid (E)-D-erythro-2-amino-1-(dihydrogenphosphate)-4-octadecene-1,3-diol C18H38NO5P D07UGA LS-185514 [ Show all ] |
| Inchi Key | DUYSYHSSBDVJSM-KRWOKUGFSA-N |
| Inchi ID | InChI=1S/C18H38NO5P/c1-2-3-4-5-6-7-8-9-10-11-12-13-14-15-18(20)17(19)16-24-25(21,22)23/h14-15,17-18,20H,2-13,16,19H2,1H3,(H2,21,22,23)/b15-14+/t17-,18+/m0/s1 |
| PubChem CID | 5283560 |
| ChEMBL | CHEMBL225155 |
| IUPHAR | 911 |
| BindingDB | 50158348 |
| DrugBank | N/A |
Structure | 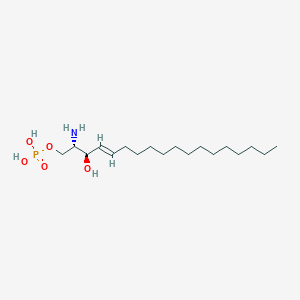 |
| Lipinski's druglikeness | This ligand satisfies Lipinski's rule of five. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| EC50 | 0.158489 - 3.98108 nM | PMID11705398, PMID17114004, PMID11967257 | IUPHAR |
| EC50 | 0.2 nM | PMID20446681 | ChEMBL |
| EC50 | 0.2 nM | PMID20446681 | BindingDB |
| EC50 | 0.449 nM | PMID20188554 | BindingDB,ChEMBL |
| EC50 | 0.7 nM | PMID20446681 | BindingDB |
| EC50 | 0.7 nM | PMID20446681 | ChEMBL |
| EC50 | 1.1 nM | PMID14505636 | BindingDB,ChEMBL |
| EC50 | 1.2 nM | PMID15341948 | BindingDB,ChEMBL |
| EC50 | 1.6 nM | PMID17070046 | BindingDB,ChEMBL |
| EC50 | 2.4 nM | PMID20304639, PMID23245510 | BindingDB,ChEMBL |
| EC50 | 3.0 nM | PMID19081720 | BindingDB,ChEMBL |
| EC50 | 3.94 nM | PMID24900589 | ChEMBL |
| EC50 | 8.7 nM | PMID15982878, PMID14505636 | BindingDB,ChEMBL |
| EC50 | 8.7 nM | PMID15982878 | BindingDB |
| EC50 | 79.0 nM | PMID26687487 | BindingDB,ChEMBL |
| Emax | 1.0 - | PMID15982878 | ChEMBL |
| IC50 | 0.04 nM | PMID15149705 | ChEMBL |
| IC50 | 0.04 nM | PMID15149705 | BindingDB |
| IC50 | 0.26 nM | PMID15615513 | BindingDB,ChEMBL |
| IC50 | 0.66 nM | PMID19081720 | BindingDB,ChEMBL |
| IC50 | 0.92 nM | PMID20304639 | ChEMBL |
| Kd | 0.23 - 26.2 nM | PMID10446161, PMID9988698, PMID17170199 | IUPHAR |
zhanglab![]() zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218