

You can:
| Name | Adenosine receptor A3 |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | ADORA3 |
| Synonym | ARA3 Adenosine receptor A3 A3AR A3 receptor TGPCR1 |
| Disease | Cerebrovascular ischaemia Malaria Ischemia Inflammation Hepatocellular carcinoma; Hepatitis C virus infection [ Show all ] |
| Length | 318 |
| Amino acid sequence | MPNNSTALSLANVTYITMEIFIGLCAIVGNVLVICVVKLNPSLQTTTFYFIVSLALADIAVGVLVMPLAIVVSLGITIHFYSCLFMTCLLLIFTHASIMSLLAIAVDRYLRVKLTVRYKRVTTHRRIWLALGLCWLVSFLVGLTPMFGWNMKLTSEYHRNVTFLSCQFVSVMRMDYMVYFSFLTWIFIPLVVMCAIYLDIFYIIRNKLSLNLSNSKETGAFYGREFKTAKSLFLVLFLFALSWLPLSIINCIIYFNGEVPQLVLYMGILLSHANSMMNPIVYAYKIKKFKETYLLILKACVVCHPSDSLDTSIEKNSE |
| UniProt | P0DMS8 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | N/A |
| 3D structure model | No available structures or models |
| BioLiP | N/A |
| Therapeutic Target Database | T36059 |
| ChEMBL | CHEMBL256 |
| IUPHAR | 21 |
| DrugBank | BE0000354 |
| Name | 8-Cyclopentyl-1,3-dipropylxanthine |
|---|---|
| Molecular formula | C16H24N4O2 |
| IUPAC name | 8-cyclopentyl-1,3-dipropyl-7H-purine-2,6-dione |
| Molecular weight | 304.394 |
| Hydrogen bond acceptor | 3 |
| Hydrogen bond donor | 1 |
| XlogP | 4.0 |
| Synonyms | FFBDFADSZUINTG-UHFFFAOYSA-N ZX-AT022179 HMS500I19 KBio3_001906 LS-126534 [ Show all ] |
| Inchi Key | FFBDFADSZUINTG-UHFFFAOYSA-N |
| Inchi ID | InChI=1S/C16H24N4O2/c1-3-9-19-14-12(15(21)20(10-4-2)16(19)22)17-13(18-14)11-7-5-6-8-11/h11H,3-10H2,1-2H3,(H,17,18) |
| PubChem CID | 1329 |
| ChEMBL | CHEMBL183 |
| IUPHAR | 386 |
| BindingDB | 21173 |
| DrugBank | N/A |
Structure | 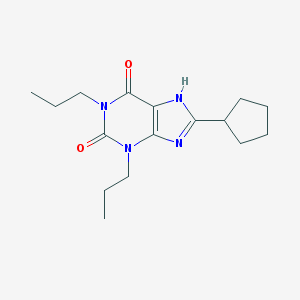 |
| Lipinski's druglikeness | This ligand satisfies Lipinski's rule of five. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| Ki | <1000.0 nM | PMID17927167, PMID16366607, PMID24077183, PMID18269230 | BindingDB,ChEMBL |
| Ki | <10000.0 nM | PMID27282729, PMID22257095, PMID11462973 | BindingDB,ChEMBL |
| Ki | 200.0 nM | PMID9767636 | BindingDB,ChEMBL |
| Ki | 243.0 nM | PMID20188574 | PDSP,BindingDB,ChEMBL |
| Ki | 243.0 - 3960.0 nM | PMID8234299, PMID9459566, PMID10779381, PMID16902942, PMID11906291, PMID9351976 | IUPHAR |
| Ki | 509.0 nM | PMID11809867 | PDSP,BindingDB |
| Ki | 758.57 nM | PMID8234299 | PDSP,BindingDB |
| Ki | 759.0 nM | PMID15771453, PMID9767636 | BindingDB,ChEMBL |
| Ki | 795.0 nM | PMID15771453, PMID11906291 | PDSP,BindingDB,ChEMBL |
| Ki | 1000.0 nM | PMID11809867 | PDSP,BindingDB |
| Ki | 1200.0 nM | PMID10779381 | PDSP,BindingDB |
| Ki | 1210.0 nM | PMID10779381 | PDSP,BindingDB |
| Ki | 1300.0 nM | PMID19301821, PMID20937560, PMID17665891, PMID15214785, PMID16789747, PMID10956220, PMID18468446, PMID16335918 | BindingDB,ChEMBL |
| Ki | 1320.0 nM | PMID23200243 | ChEMBL |
| Ki | 1700.0 nM | PMID18258439, PMID18637670, PMID15771447, PMID9767636 | BindingDB,ChEMBL |
| Ki | 1722.0 nM | PMID26824742 | BindingDB,ChEMBL |
| Ki | 3960.0 nM | PMID9459566 | PDSP,BindingDB |
| Ki | 3980.0 nM | PMID23200243 | ChEMBL |
| Ki | 4000.0 nM | PMID20408530, PMID19282184 | BindingDB,ChEMBL |
zhanglab![]() zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218