

You can:
| Name | 5-hydroxytryptamine receptor 2B |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | HTR2B |
| Synonym | stomach fundus serotonin receptor serotonin receptor 2B 5-HT2F 5-HT-2B 5-HT2B [ Show all ] |
| Disease | Psychoses Migraine Irritable bowel syndrome Depression; Cerebral infarction Coronary heart disease [ Show all ] |
| Length | 481 |
| Amino acid sequence | MALSYRVSELQSTIPEHILQSTFVHVISSNWSGLQTESIPEEMKQIVEEQGNKLHWAALLILMVIIPTIGGNTLVILAVSLEKKLQYATNYFLMSLAVADLLVGLFVMPIALLTIMFEAMWPLPLVLCPAWLFLDVLFSTASIMHLCAISVDRYIAIKKPIQANQYNSRATAFIKITVVWLISIGIAIPVPIKGIETDVDNPNNITCVLTKERFGDFMLFGSLAAFFTPLAIMIVTYFLTIHALQKKAYLVKNKPPQRLTWLTVSTVFQRDETPCSSPEKVAMLDGSRKDKALPNSGDETLMRRTSTIGKKSVQTISNEQRASKVLGIVFFLFLLMWCPFFITNITLVLCDSCNQTTLQMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAFGRYITCNYRATKSVKTLRKRSSKIYFRNPMAENSKFFKKHGIRNGINPAMYQSPMRLRSSTIQSSSIILLDTLLLTENEGDKTEEQVSYV |
| UniProt | P41595 |
| Protein Data Bank | 5tud, 5tvn, 6drx, 4ib4, 4nc3, 6dry, 6drz, 6ds0 |
| GPCR-HGmod model | P41595 |
| 3D structure model | This structure is from PDB ID 5tud. |
| BioLiP | BL0425280, BL0425283, BL0425282, BL0239858, BL0239859, BL0265182, BL0265183, BL0368464, BL0425281, BL0385686,BL0385687, BL0425284, BL0425285, BL0425286, BL0368465, BL0425287 |
| Therapeutic Target Database | T31204 |
| ChEMBL | CHEMBL1833 |
| IUPHAR | 7 |
| DrugBank | BE0000393 |
| Name | Ketanserin |
|---|---|
| Molecular formula | C22H22FN3O3 |
| IUPAC name | 3-[2-[4-(4-fluorobenzoyl)piperidin-1-yl]ethyl]-1H-quinazoline-2,4-dione |
| Molecular weight | 395.434 |
| Hydrogen bond acceptor | 5 |
| Hydrogen bond donor | 1 |
| XlogP | 2.6 |
| Synonyms | R-41468 CTK8G0431 Serefrex DTXSID3023188 GTPL88 [ Show all ] |
| Inchi Key | FPCCSQOGAWCVBH-UHFFFAOYSA-N |
| Inchi ID | InChI=1S/C22H22FN3O3/c23-17-7-5-15(6-8-17)20(27)16-9-11-25(12-10-16)13-14-26-21(28)18-3-1-2-4-19(18)24-22(26)29/h1-8,16H,9-14H2,(H,24,29) |
| PubChem CID | 3822 |
| ChEMBL | CHEMBL51 |
| IUPHAR | 88, 197 |
| BindingDB | 21395 |
| DrugBank | DB12465 |
Structure | 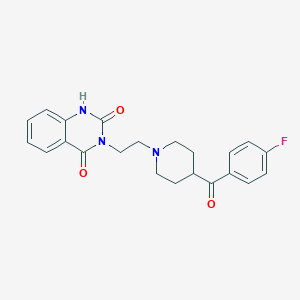 |
| Lipinski's druglikeness | This ligand satisfies Lipinski's rule of five. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| IC50 | 290.0 nM | PMID23403082 | ChEMBL |
| Inhibition | 42.0 % | PMID23403082 | ChEMBL |
| Ki | 180.0 nM | PMID23403082 | ChEMBL |
| Ki | 199.526 - 794.328 nM | PMID15322733, PMID8845011, PMID12738034, PMID8078486, PMID9459568 | IUPHAR |
| Ki | 217.0 nM | PMID9459568 | PDSP,BindingDB |
| Ki | 234.42 nM | PMID11882920 | BindingDB |
| Ki | 234.423 nM | PMID11882920 | PDSP |
| Ki | 372.0 nM | PMID9459568, PMID8845011 | PDSP,BindingDB |
| Ki | 376.0 nM | PMID8078486 | PDSP,BindingDB |
| Ki | 395.0 nM | PMID8632342 | PDSP,BindingDB |
| Ki | 630.95 nM | PMID7582481 | PDSP,BindingDB |
| Ki | 630.957 nM | PMID10821800 | PDSP |
| Ki | 741.31 nM | PMID15322733 | BindingDB |
| Ki | 741.31 nM | PMID15322733 | PDSP |
| Ki | 3235.93 nM | PMID9225287 | PDSP,BindingDB |
zhanglab![]() zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218