

You can:
| Name | Motilin receptor |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | MLNR |
| Synonym | GPR38 G protein-coupled receptor 38 G-protein coupled receptor 38 MTLR1 MTLR [ Show all ] |
| Disease | Gastrointestinal disorders; Gastroesophageal reflux disease Irritable bowel syndrome Gastrointestinal disease Gastroesophageal reflux disease Gastric motility disorder [ Show all ] |
| Length | 412 |
| Amino acid sequence | MGSPWNGSDGPEGAREPPWPALPPCDERRCSPFPLGALVPVTAVCLCLFVVGVSGNVVTVMLIGRYRDMRTTTNLYLGSMAVSDLLILLGLPFDLYRLWRSRPWVFGPLLCRLSLYVGEGCTYATLLHMTALSVERYLAICRPLRARVLVTRRRVRALIAVLWAVALLSAGPFLFLVGVEQDPGISVVPGLNGTARIASSPLASSPPLWLSRAPPPSPPSGPETAEAAALFSRECRPSPAQLGALRVMLWVTTAYFFLPFLCLSILYGLIGRELWSSRRPLRGPAASGRERGHRQTVRVLLVVVLAFIICWLPFHVGRIIYINTEDSRMMYFSQYFNIVALQLFYLSASINPILYNLISKKYRAAAFKLLLARKSRPRGFHRSRDTAGEVAGDTGGDTVGYTETSANVKTMG |
| UniProt | O43193 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | O43193 |
| 3D structure model | This predicted structure model is from GPCR-EXP O43193. |
| BioLiP | N/A |
| Therapeutic Target Database | T62306 |
| ChEMBL | CHEMBL2203 |
| IUPHAR | 297 |
| DrugBank | BE0003521 |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 917 | 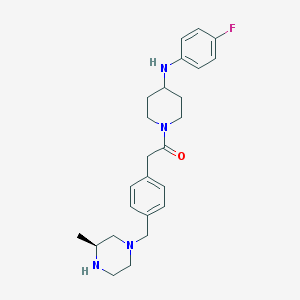 CHEMBL489094 CHEMBL489094 | C25H33FN4O | 424.564 | 5 / 2 | 3.3 | Yes |
| 1228 | 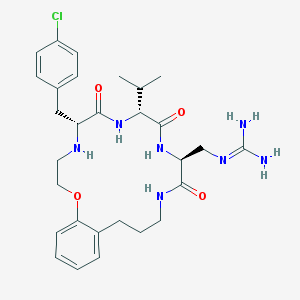 CHEMBL232154 CHEMBL232154 | C29H40ClN7O4 | 586.134 | 6 / 6 | 2.4 | No |
| 3430 | 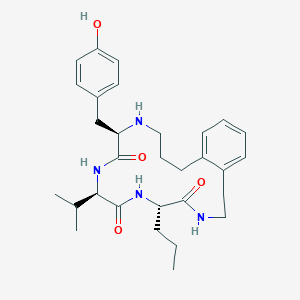 CHEMBL437156 CHEMBL437156 | C30H42N4O4 | 522.69 | 5 / 5 | 4.5 | No |
| 3969 | 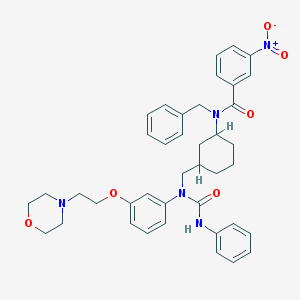 CHEMBL210899 CHEMBL210899 | C40H45N5O6 | 691.829 | 7 / 1 | 6.3 | No |
| 4019 | 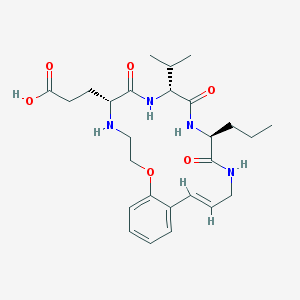 CHEMBL215292 CHEMBL215292 | C26H38N4O6 | 502.612 | 7 / 5 | 0.1 | No |
| 8140 | 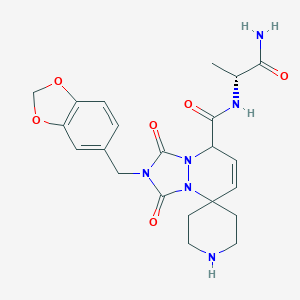 CHEMBL44542 CHEMBL44542 | C22H26N6O6 | 470.486 | 7 / 3 | -0.2 | Yes |
| 8141 | 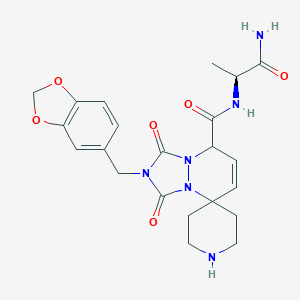 CHEMBL296962 CHEMBL296962 | C22H26N6O6 | 470.486 | 7 / 3 | -0.2 | Yes |
| 553294 | 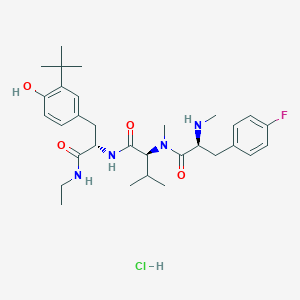 MA2029 MA2029 | C31H46ClFN4O4 | 593.181 | 6 / 5 | N/A | No |
| 10860 | 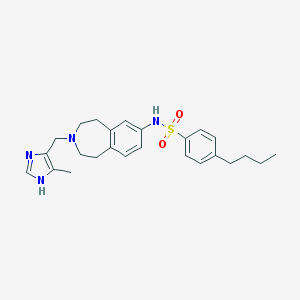 CHEMBL1079186 CHEMBL1079186 | C25H32N4O2S | 452.617 | 5 / 2 | 4.7 | Yes |
| 12209 | 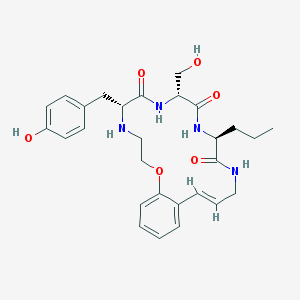 CHEMBL214543 CHEMBL214543 | C28H36N4O6 | 524.618 | 7 / 6 | 2.7 | No |
| 13474 | 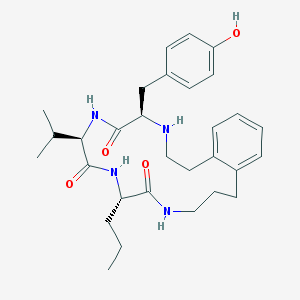 CHEMBL214815 CHEMBL214815 | C30H42N4O4 | 522.69 | 5 / 5 | 4.5 | No |
| 16586 | 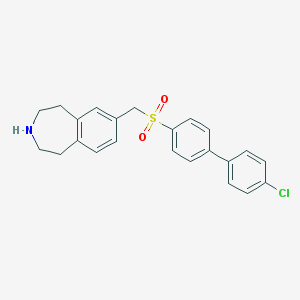 CHEMBL1078344 CHEMBL1078344 | C23H22ClNO2S | 411.944 | 3 / 1 | 4.8 | Yes |
| 16970 | 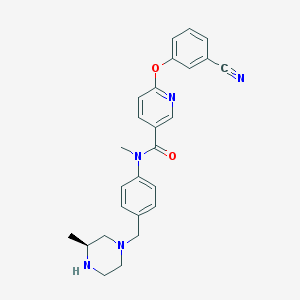 SCHEMBL2768193 SCHEMBL2768193 | C26H27N5O2 | 441.535 | 6 / 1 | 3.1 | Yes |
| 17611 | 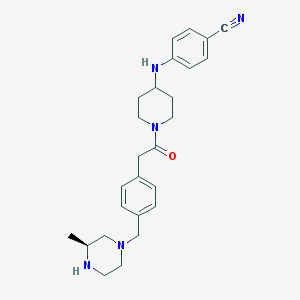 SCHEMBL1704096 SCHEMBL1704096 | C26H33N5O | 431.584 | 5 / 2 | 2.9 | Yes |
| 17612 | 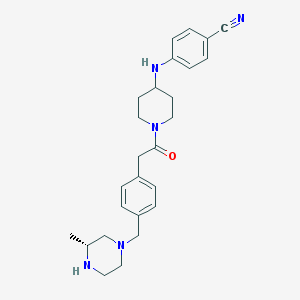 SCHEMBL1704626 SCHEMBL1704626 | C26H33N5O | 431.584 | 5 / 2 | 2.9 | Yes |
| 21041 | 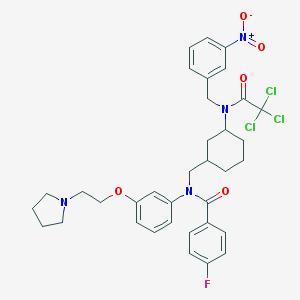 CHEMBL378317 CHEMBL378317 | C35H38Cl3FN4O5 | 720.06 | 7 / 0 | 7.7 | No |
| 21684 | 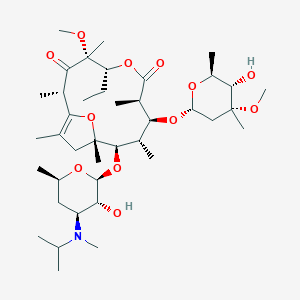 GM611 GM611 | C40H69NO12 | 755.987 | 13 / 2 | 4.1 | No |
| 548134 | 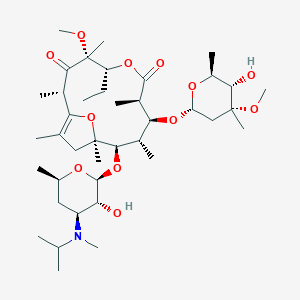 mitemcinal mitemcinal | C40H69NO12 | 755.987 | 13 / 2 | 4.1 | No |
| 24775 | 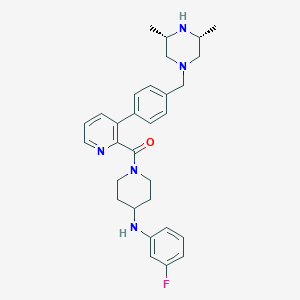 CHEMBL463881 CHEMBL463881 | C30H36FN5O | 501.65 | 6 / 2 | 4.7 | No |
| 25952 | 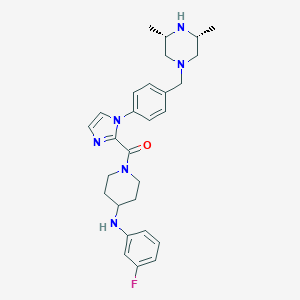 CHEMBL480219 CHEMBL480219 | C28H35FN6O | 490.627 | 6 / 2 | 3.8 | Yes |
| 27462 | 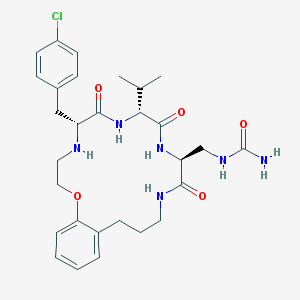 CHEMBL232359 CHEMBL232359 | C29H39ClN6O5 | 587.118 | 6 / 6 | 3.2 | No |
| 27760 | 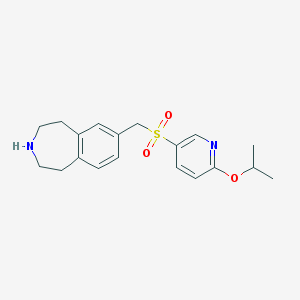 CHEMBL1078461 CHEMBL1078461 | C19H24N2O3S | 360.472 | 5 / 1 | 2.6 | Yes |
| 28094 | 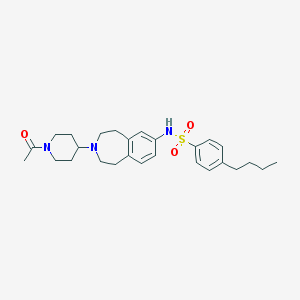 CHEMBL1078863 CHEMBL1078863 | C27H37N3O3S | 483.671 | 5 / 1 | 4.7 | Yes |
| 28170 | 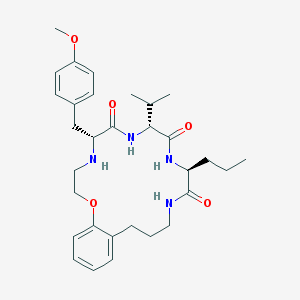 CHEMBL384501 CHEMBL384501 | C31H44N4O5 | 552.716 | 6 / 4 | 4.5 | No |
| 28944 | 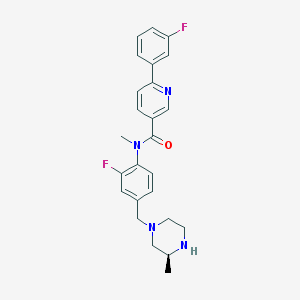 SCHEMBL2767377 SCHEMBL2767377 | C25H26F2N4O | 436.507 | 6 / 1 | 3.3 | Yes |
| 34126 | 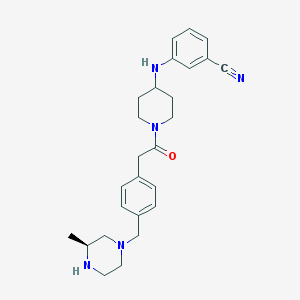 CHEMBL489480 CHEMBL489480 | C26H33N5O | 431.584 | 5 / 2 | 2.9 | Yes |
| 34127 | 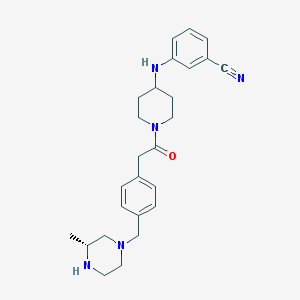 SCHEMBL1704116 SCHEMBL1704116 | C26H33N5O | 431.584 | 5 / 2 | 2.9 | Yes |
| 40367 | 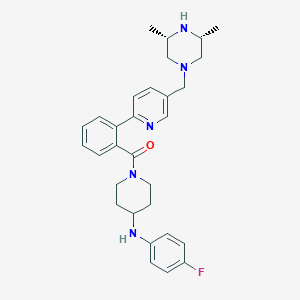 CHEMBL481001 CHEMBL481001 | C30H36FN5O | 501.65 | 6 / 2 | 4.4 | No |
| 43386 | 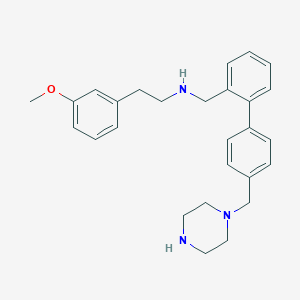 CHEMBL475808 CHEMBL475808 | C27H33N3O | 415.581 | 4 / 2 | 4.0 | Yes |
| 43928 | 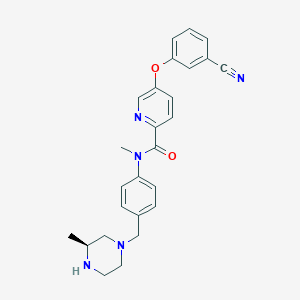 SCHEMBL2766955 SCHEMBL2766955 | C26H27N5O2 | 441.535 | 6 / 1 | 3.1 | Yes |
| 44203 | 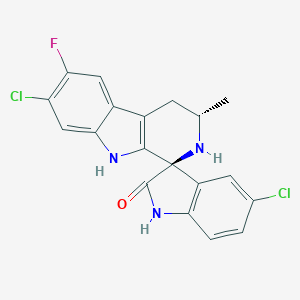 Cipargamin Cipargamin | C19H14Cl2FN3O | 390.239 | 3 / 3 | 3.9 | Yes |
| 46465 | 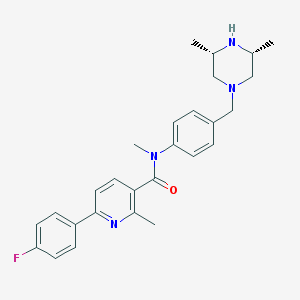 SCHEMBL2765432 SCHEMBL2765432 | C27H31FN4O | 446.57 | 5 / 1 | 4.1 | Yes |
| 48585 |  CHEMBL386036 CHEMBL386036 | C28H36N4O6 | 524.618 | 7 / 6 | 2.8 | No |
| 48586 |  CHEMBL215440 CHEMBL215440 | C30H42N4O4 | 522.69 | 5 / 4 | 4.5 | No |
| 50250 |  SCHEMBL2765776 SCHEMBL2765776 | C26H27N5O | 425.536 | 5 / 1 | 3.2 | Yes |
| 51575 |  CHEMBL262052 CHEMBL262052 | C39H69NO12 | 743.976 | 13 / 3 | 3.3 | No |
| 52581 | 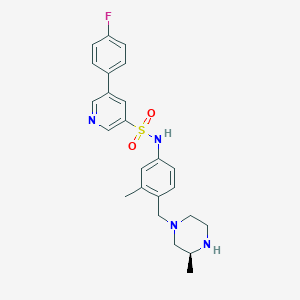 SCHEMBL2004331 SCHEMBL2004331 | C24H27FN4O2S | 454.564 | 7 / 2 | 3.1 | Yes |
| 52963 | 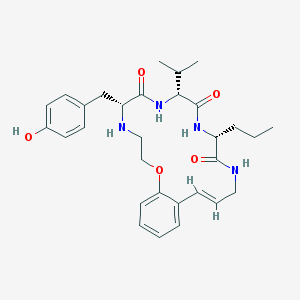 CHEMBL383881 CHEMBL383881 | C30H40N4O5 | 536.673 | 6 / 5 | 4.1 | No |
| 52964 | 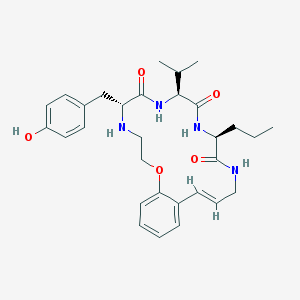 CHEMBL387446 CHEMBL387446 | C30H40N4O5 | 536.673 | 6 / 5 | 4.1 | No |
| 52965 | 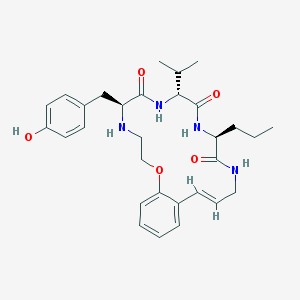 CHEMBL214702 CHEMBL214702 | C30H40N4O5 | 536.673 | 6 / 5 | 4.1 | No |
| 52966 | 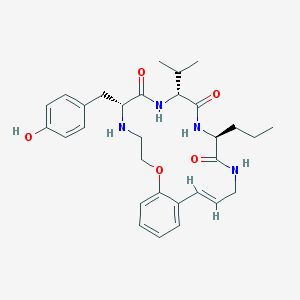 CHEMBL214935 CHEMBL214935 | C30H40N4O5 | 536.673 | 6 / 5 | 4.1 | No |
| 53419 | 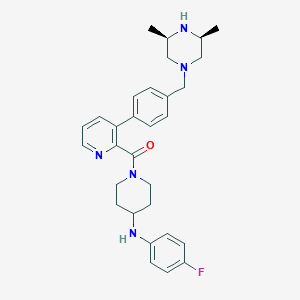 CHEMBL473921 CHEMBL473921 | C30H36FN5O | 501.65 | 6 / 2 | 4.7 | No |
| 54817 | 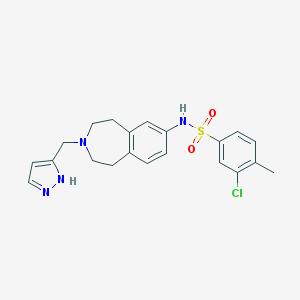 CHEMBL1079311 CHEMBL1079311 | C21H23ClN4O2S | 430.951 | 5 / 2 | 3.6 | Yes |
| 56828 | 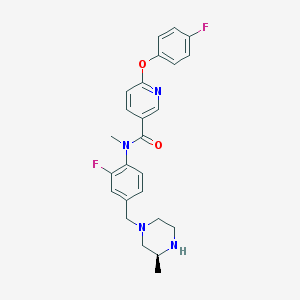 SCHEMBL2765921 SCHEMBL2765921 | C25H26F2N4O2 | 452.506 | 7 / 1 | 3.5 | Yes |
| 57677 | 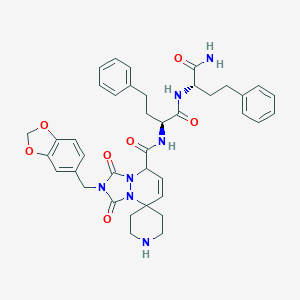 CHEMBL296748 CHEMBL296748 | C39H43N7O7 | 721.815 | 8 / 4 | 3.5 | No |
| 57736 | 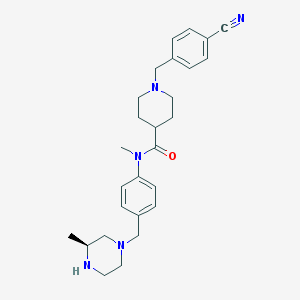 SCHEMBL2767107 SCHEMBL2767107 | C27H35N5O | 445.611 | 5 / 1 | 2.6 | Yes |
| 58748 | 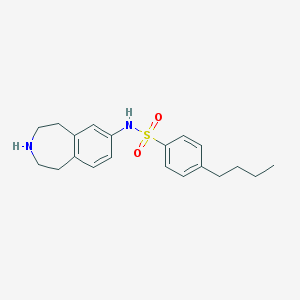 CHEMBL266018 CHEMBL266018 | C20H26N2O2S | 358.5 | 4 / 2 | 4.2 | Yes |
| 60585 | 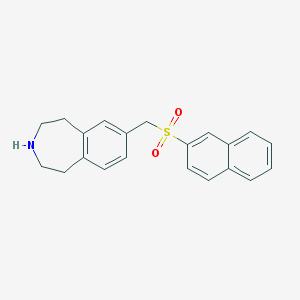 CHEMBL1078343 CHEMBL1078343 | C21H21NO2S | 351.464 | 3 / 1 | 3.8 | Yes |
| 61194 | 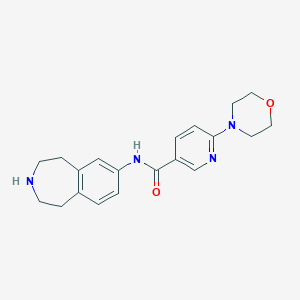 CHEMBL1078540 CHEMBL1078540 | C20H24N4O2 | 352.438 | 5 / 2 | 1.6 | Yes |
| 61508 | 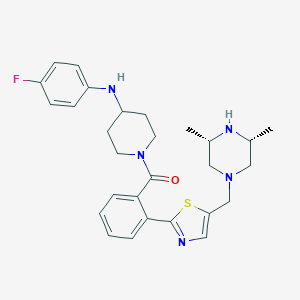 CHEMBL481192 CHEMBL481192 | C28H34FN5OS | 507.672 | 7 / 2 | 4.5 | No |
| 64731 | 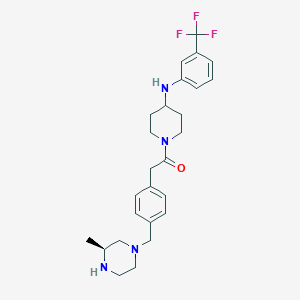 SCHEMBL1704702 SCHEMBL1704702 | C26H33F3N4O | 474.572 | 7 / 2 | 4.0 | Yes |
| 64732 | 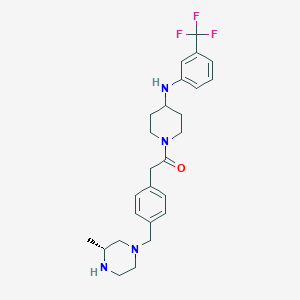 SCHEMBL1704164 SCHEMBL1704164 | C26H33F3N4O | 474.572 | 7 / 2 | 4.0 | Yes |
| 65518 | 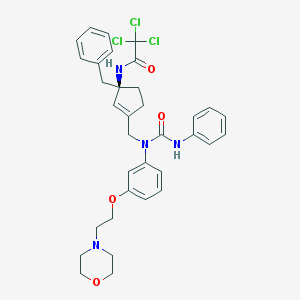 CHEMBL379519 CHEMBL379519 | C34H37Cl3N4O4 | 672.044 | 5 / 2 | 5.7 | No |
| 66992 | 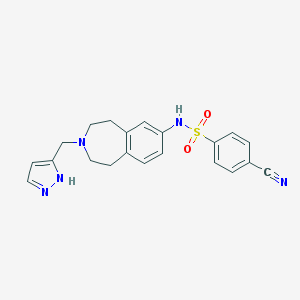 CHEMBL1081860 CHEMBL1081860 | C21H21N5O2S | 407.492 | 6 / 2 | 2.3 | Yes |
| 67141 | 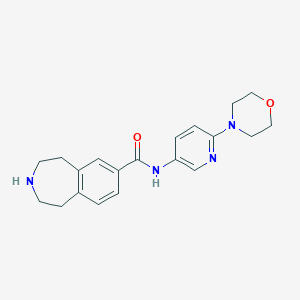 CHEMBL1078639 CHEMBL1078639 | C20H24N4O2 | 352.438 | 5 / 2 | 1.6 | Yes |
| 67756 | 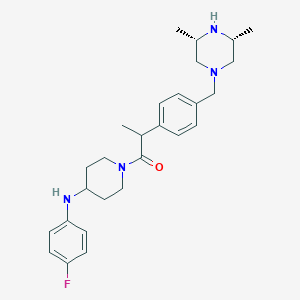 CHEMBL489297 CHEMBL489297 | C27H37FN4O | 452.618 | 5 / 2 | 4.3 | Yes |
| 69833 | 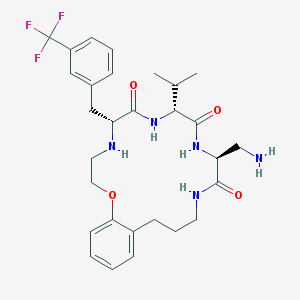 CHEMBL274876 CHEMBL274876 | C29H38F3N5O4 | 577.649 | 9 / 5 | 3.8 | No |
| 71437 | 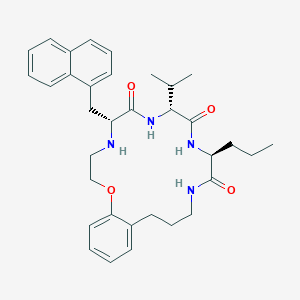 CHEMBL385087 CHEMBL385087 | C34H44N4O4 | 572.75 | 5 / 4 | 5.8 | No |
| 553632 | 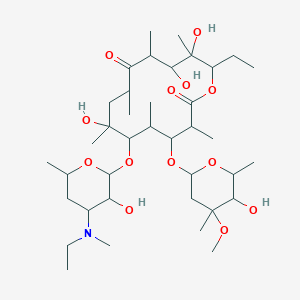 N-ethyl,N-methyl EM-A N-ethyl,N-methyl EM-A | C38H69NO13 | 747.964 | 14 / 5 | 3.1 | No |
| 74216 | 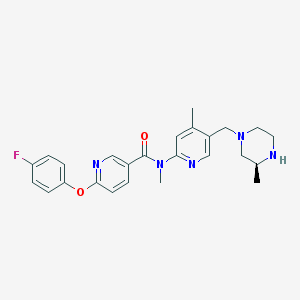 SCHEMBL2765637 SCHEMBL2765637 | C25H28FN5O2 | 449.53 | 7 / 1 | 3.1 | Yes |
| 76260 | 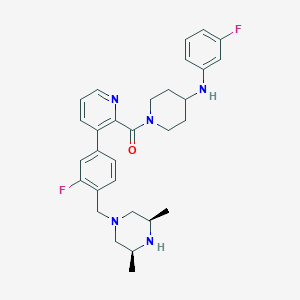 CHEMBL465094 CHEMBL465094 | C30H35F2N5O | 519.641 | 7 / 2 | 4.8 | No |
| 81803 | 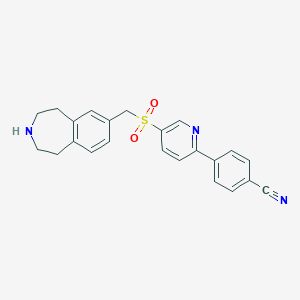 CHEMBL1078269 CHEMBL1078269 | C23H21N3O2S | 403.5 | 5 / 1 | 2.8 | Yes |
| 523953 | 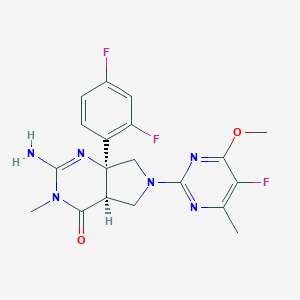 BDBM9488 BDBM9488 | C19H19F3N6O2 | 420.396 | 9 / 1 | 1.3 | Yes |
| 83477 | 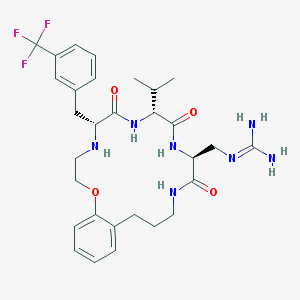 CHEMBL396413 CHEMBL396413 | C30H40F3N7O4 | 619.69 | 9 / 6 | 2.6 | No |
| 83694 | 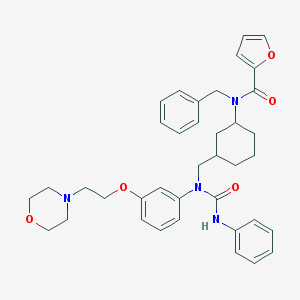 CHEMBL210923 CHEMBL210923 | C38H44N4O5 | 636.793 | 6 / 1 | 5.9 | No |
| 85503 | 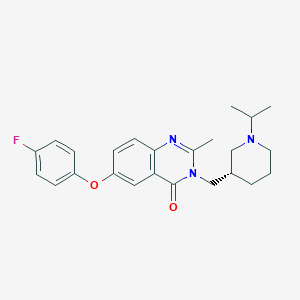 yil-781 yil-781 | C24H28FN3O2 | 409.505 | 5 / 0 | 4.1 | Yes |
| 86525 | 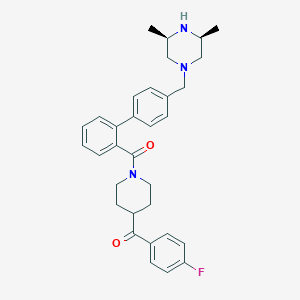 CHEMBL480210 CHEMBL480210 | C32H36FN3O2 | 513.657 | 5 / 1 | 5.0 | No |
| 89166 | 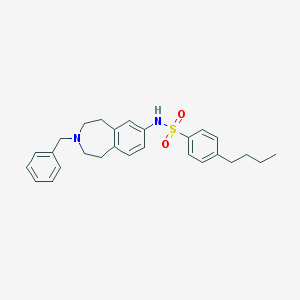 CHEMBL1078807 CHEMBL1078807 | C27H32N2O2S | 448.625 | 4 / 1 | 6.2 | No |
| 89418 | 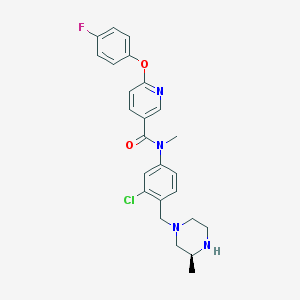 SCHEMBL2765832 SCHEMBL2765832 | C25H26ClFN4O2 | 468.957 | 6 / 1 | 4.1 | Yes |
| 90989 | 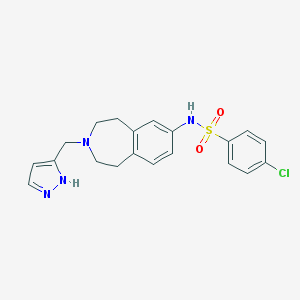 CHEMBL1078104 CHEMBL1078104 | C20H21ClN4O2S | 416.924 | 5 / 2 | 3.2 | Yes |
| 91889 | 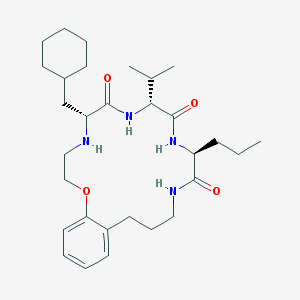 CHEMBL384204 CHEMBL384204 | C30H48N4O4 | 528.738 | 5 / 4 | 5.7 | No |
| 553723 | 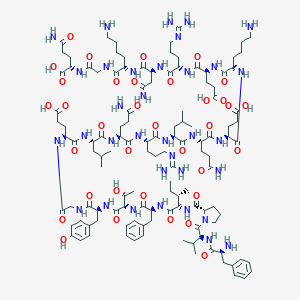 [Leu13]motilin [Leu13]motilin | C121H190N34O35 | 2681.05 | 40 / 37 | -14.1 | No |
| 96501 | 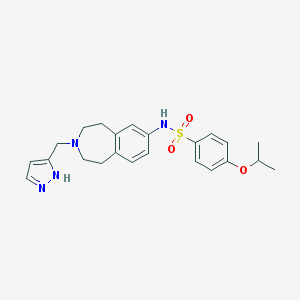 CHEMBL1078208 CHEMBL1078208 | C23H28N4O3S | 440.562 | 6 / 2 | 3.4 | Yes |
| 97145 | 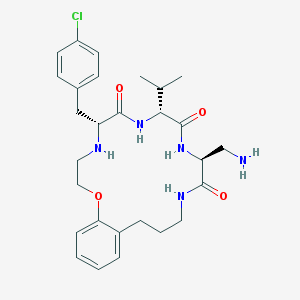 CHEMBL391045 CHEMBL391045 | C28H38ClN5O4 | 544.093 | 6 / 5 | 3.5 | No |
| 98408 | 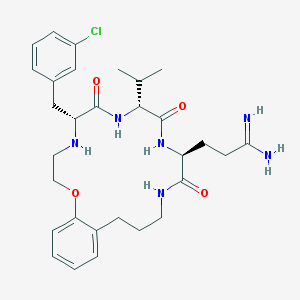 CHEMBL234810 CHEMBL234810 | C30H41ClN6O4 | 585.146 | 6 / 6 | 3.2 | No |
| 100910 | 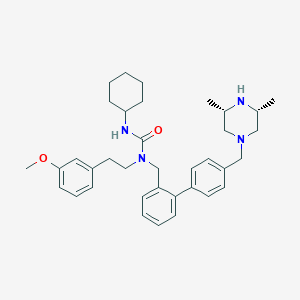 CHEMBL501315 CHEMBL501315 | C36H48N4O2 | 568.806 | 4 / 2 | 6.5 | No |
| 104151 | 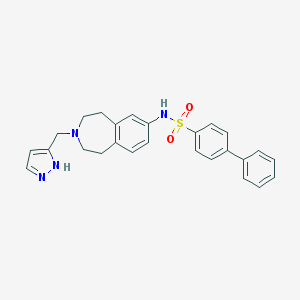 CHEMBL1078207 CHEMBL1078207 | C26H26N4O2S | 458.58 | 5 / 2 | 4.2 | Yes |
| 110187 | 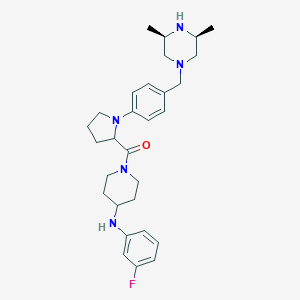 CHEMBL518261 CHEMBL518261 | C29H40FN5O | 493.671 | 6 / 2 | 4.5 | Yes |
| 111802 | 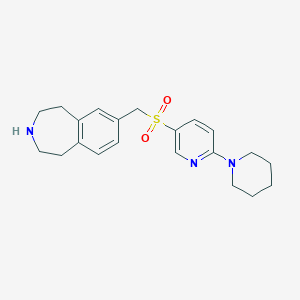 CHEMBL1078463 CHEMBL1078463 | C21H27N3O2S | 385.526 | 5 / 1 | 2.8 | Yes |
| 112980 | 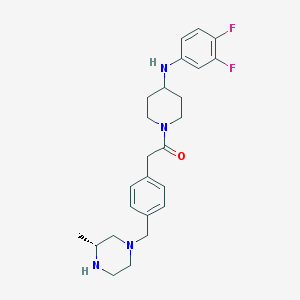 SCHEMBL1704649 SCHEMBL1704649 | C25H32F2N4O | 442.555 | 6 / 2 | 3.4 | Yes |
| 112981 | 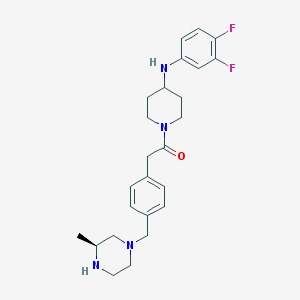 SCHEMBL1704133 SCHEMBL1704133 | C25H32F2N4O | 442.555 | 6 / 2 | 3.4 | Yes |
| 113053 | 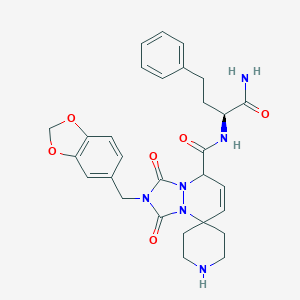 CHEMBL45905 CHEMBL45905 | C29H32N6O6 | 560.611 | 7 / 3 | 1.8 | No |
| 113054 | 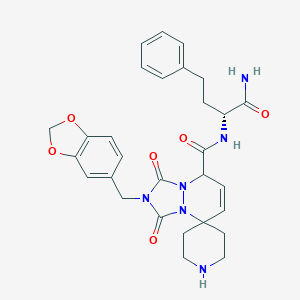 CHEMBL33827 CHEMBL33827 | C29H32N6O6 | 560.611 | 7 / 3 | 1.8 | No |
| 117778 | 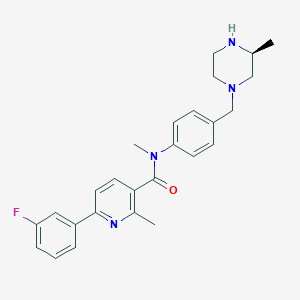 SCHEMBL2768052 SCHEMBL2768052 | C26H29FN4O | 432.543 | 5 / 1 | 3.6 | Yes |
| 120344 | 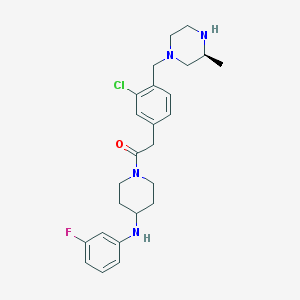 SCHEMBL1704149 SCHEMBL1704149 | C25H32ClFN4O | 459.006 | 5 / 2 | 3.9 | Yes |
| 120572 | 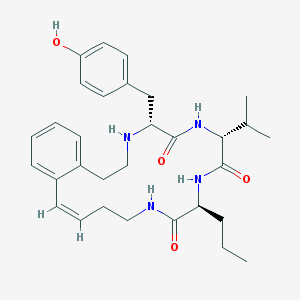 CHEMBL384698 CHEMBL384698 | C31H42N4O4 | 534.701 | 5 / 5 | 4.9 | No |
| 121520 | 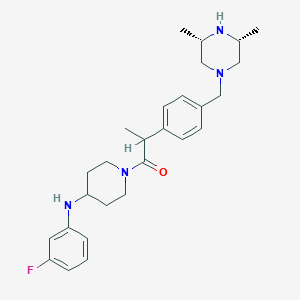 SCHEMBL1704176 SCHEMBL1704176 | C27H37FN4O | 452.618 | 5 / 2 | 4.3 | Yes |
| 124775 | 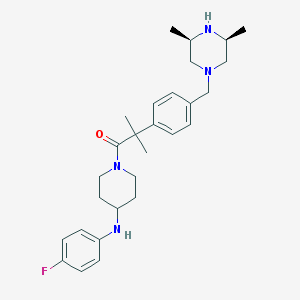 CHEMBL489479 CHEMBL489479 | C28H39FN4O | 466.645 | 5 / 2 | 4.6 | Yes |
| 126548 | 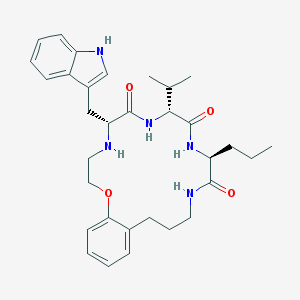 CHEMBL385673 CHEMBL385673 | C32H43N5O4 | 561.727 | 5 / 5 | 4.7 | No |
| 126786 | 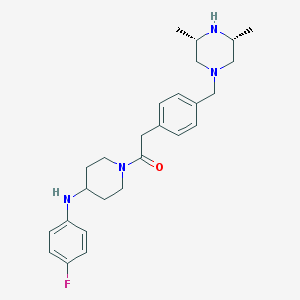 CHEMBL474934 CHEMBL474934 | C26H35FN4O | 438.591 | 5 / 2 | 3.7 | Yes |
| 129440 | 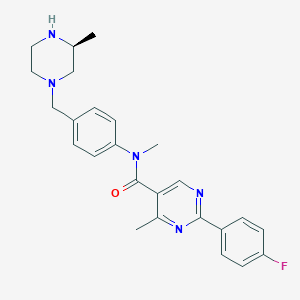 SCHEMBL2767434 SCHEMBL2767434 | C25H28FN5O | 433.531 | 6 / 1 | 3.0 | Yes |
| 129855 | 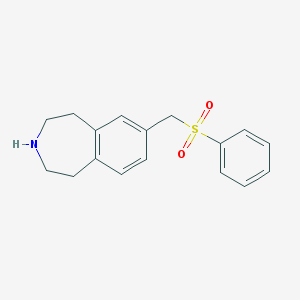 CHEMBL1077578 CHEMBL1077578 | C17H19NO2S | 301.404 | 3 / 1 | 2.5 | Yes |
| 131175 | 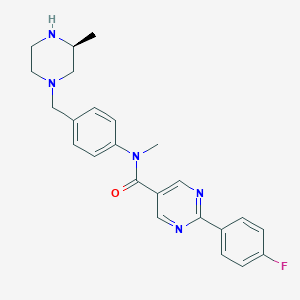 SCHEMBL2767740 SCHEMBL2767740 | C24H26FN5O | 419.504 | 6 / 1 | 2.6 | Yes |
| 131593 | 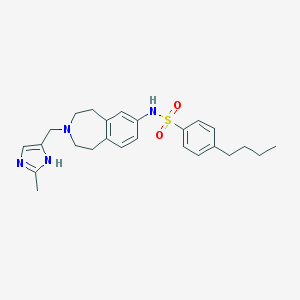 CHEMBL1079187 CHEMBL1079187 | C25H32N4O2S | 452.617 | 5 / 2 | 4.7 | Yes |
| 131994 | 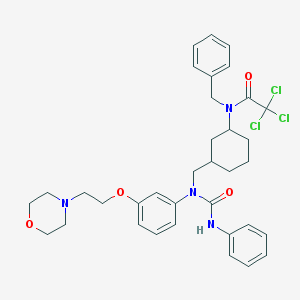 CHEMBL212044 CHEMBL212044 | C35H41Cl3N4O4 | 688.087 | 5 / 1 | 6.7 | No |
| 133534 | 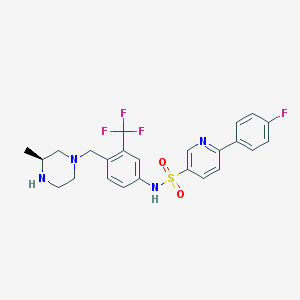 SCHEMBL2000727 SCHEMBL2000727 | C24H24F4N4O2S | 508.536 | 10 / 2 | 3.6 | No |
| 136834 | 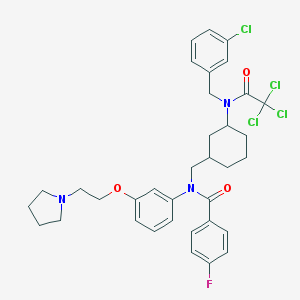 CHEMBL382612 CHEMBL382612 | C35H38Cl4FN3O3 | 709.505 | 5 / 0 | 8.5 | No |
| 138405 | 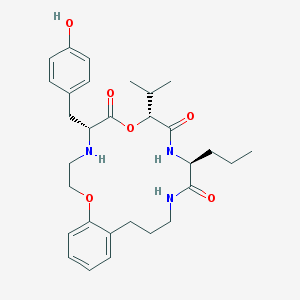 CHEMBL386626 CHEMBL386626 | C30H41N3O6 | 539.673 | 7 / 4 | 4.8 | No |
| 140727 | 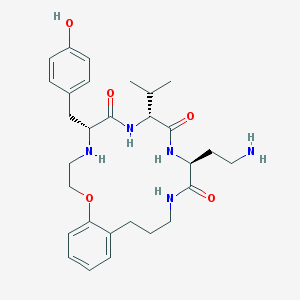 CHEMBL232342 CHEMBL232342 | C29H41N5O5 | 539.677 | 7 / 6 | 2.3 | No |
| 141661 | 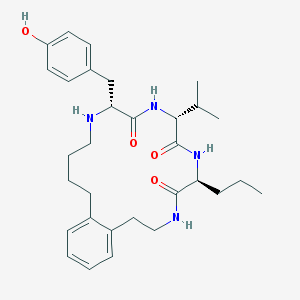 CHEMBL215963 CHEMBL215963 | C31H44N4O4 | 536.717 | 5 / 5 | 4.8 | No |
zhanglab![]() zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218