

You can:
| Name | Sphingosine 1-phosphate receptor 2 |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | S1PR2 |
| Synonym | Endothelial differentiation G-protein coupled receptor 5 Sphingosine 1-phosphate receptor Edg-5 S1P2 receptor S1P2 S1P receptor Edg-5 [ Show all ] |
| Disease | Hypertension |
| Length | 353 |
| Amino acid sequence | MGSLYSEYLNPNKVQEHYNYTKETLETQETTSRQVASAFIVILCCAIVVENLLVLIAVARNSKFHSAMYLFLGNLAASDLLAGVAFVANTLLSGSVTLRLTPVQWFAREGSAFITLSASVFSLLAIAIERHVAIAKVKLYGSDKSCRMLLLIGASWLISLVLGGLPILGWNCLGHLEACSTVLPLYAKHYVLCVVTIFSIILLAIVALYVRIYCVVRSSHADMAAPQTLALLKTVTIVLGVFIVCWLPAFSILLLDYACPVHSCPILYKAHYFFAVSTLNSLLNPVIYTWRSRDLRREVLRPLQCWRPGVGVQGRRRGGTPGHHLLPLRSSSSLERGMHMPTSPTFLEGNTVV |
| UniProt | O95136 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | O95136 |
| 3D structure model | This predicted structure model is from GPCR-EXP O95136. |
| BioLiP | N/A |
| Therapeutic Target Database | T47888 |
| ChEMBL | CHEMBL2955 |
| IUPHAR | 276 |
| DrugBank | N/A |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 521439 | 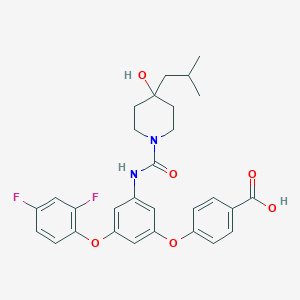 CHEMBL3759551 CHEMBL3759551 | C29H30F2N2O6 | 540.564 | 8 / 3 | 5.3 | No |
| 33 | 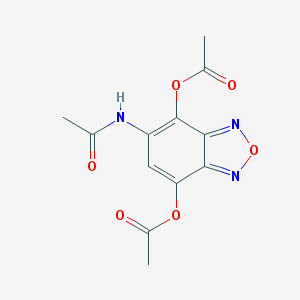 MLS000077136 MLS000077136 | C12H11N3O6 | 293.235 | 8 / 1 | -0.1 | Yes |
| 148 | 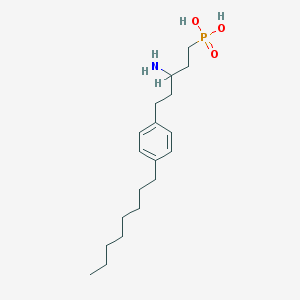 CHEMBL325198 CHEMBL325198 | C19H34NO3P | 355.459 | 4 / 3 | 2.0 | Yes |
| 1374 | 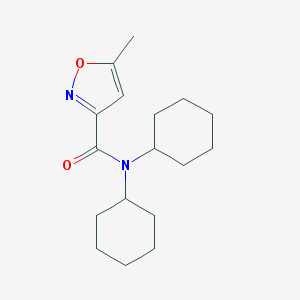 SMR000023524 SMR000023524 | C17H26N2O2 | 290.407 | 3 / 0 | 4.2 | Yes |
| 1954 |  CHEMBL2018573 CHEMBL2018573 | C28H25F2N5O3 | 517.537 | 7 / 1 | 4.9 | No |
| 2170 |  SR-02000000242 SR-02000000242 | C19H15Cl2NO2 | 360.234 | 2 / 1 | 5.8 | No |
| 441831 |  CHEMBL3400190 CHEMBL3400190 | C24H32N2O3 | 396.531 | 3 / 2 | 5.1 | No |
| 3497 |  CHEMBL377828 CHEMBL377828 | C26H29N3O3 | 431.536 | 6 / 2 | 3.0 | Yes |
| 3582 |  MLS000557830 MLS000557830 | C24H23N3O | 369.468 | 4 / 0 | 4.1 | Yes |
| 3711 |  AC1NW0NQ AC1NW0NQ | C14H13NO4S | 291.321 | 5 / 0 | 2.5 | Yes |
| 521568 |  CHEMBL3759532 CHEMBL3759532 | C32H35FN2O6 | 562.638 | 7 / 3 | 5.8 | No |
| 4386 |  CHEMBL117973 CHEMBL117973 | C18H40NO3P | 349.496 | 4 / 3 | 3.3 | Yes |
| 4625 |  SMR000019512 SMR000019512 | C15H8ClNO3S2 | 349.803 | 5 / 0 | 3.9 | Yes |
| 4935 | 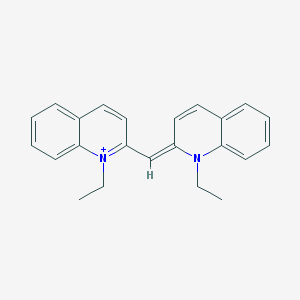 Pseudoisocyanine Pseudoisocyanine | C23H23N2+ | 327.451 | 1 / 0 | 5.6 | No |
| 4963 | 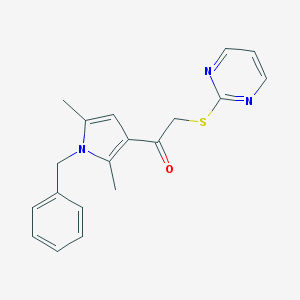 SR-01000051804-2 SR-01000051804-2 | C19H19N3OS | 337.441 | 4 / 0 | 3.6 | Yes |
| 5088 | 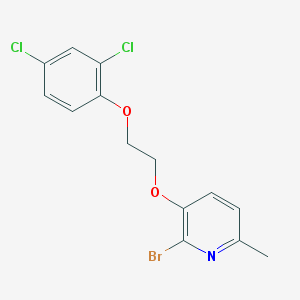 CYM50199 CYM50199 | C14H12BrCl2NO2 | 377.059 | 3 / 0 | 5.2 | No |
| 5483 | 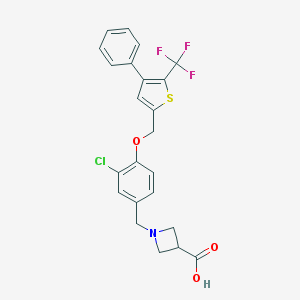 CHEMBL224571 CHEMBL224571 | C23H19ClF3NO3S | 481.914 | 8 / 1 | 3.1 | Yes |
| 5795 | 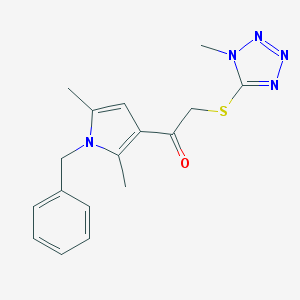 SR-01000759072-1 SR-01000759072-1 | C17H19N5OS | 341.433 | 5 / 0 | 3.0 | Yes |
| 6040 | 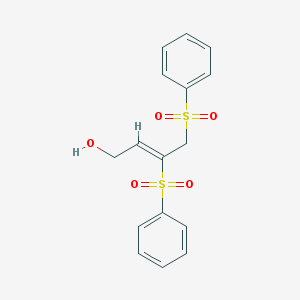 MLS000080860 MLS000080860 | C16H16O5S2 | 352.419 | 5 / 1 | 1.4 | Yes |
| 7680 | 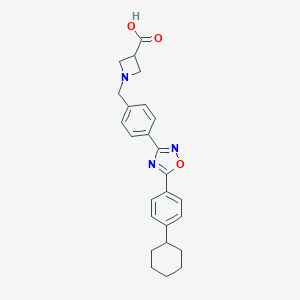 CHEMBL196534 CHEMBL196534 | C25H27N3O3 | 417.509 | 6 / 1 | 2.9 | Yes |
| 8956 | 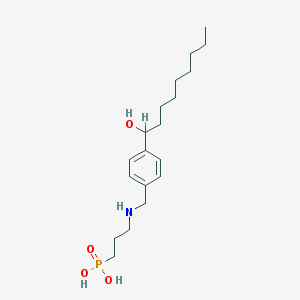 CHEMBL117007 CHEMBL117007 | C19H34NO4P | 371.458 | 5 / 4 | 0.6 | Yes |
| 10100 | 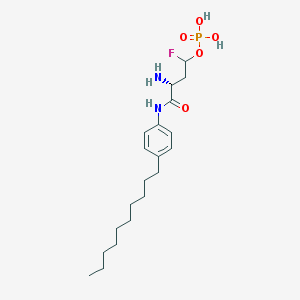 CHEMBL226612 CHEMBL226612 | C20H34FN2O5P | 432.473 | 7 / 4 | 2.2 | Yes |
| 10697 | 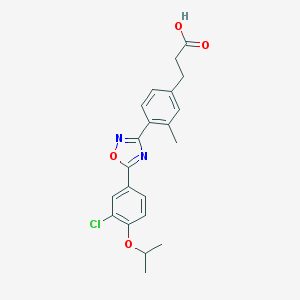 CHEMBL206940 CHEMBL206940 | C21H21ClN2O4 | 400.859 | 6 / 1 | 5.0 | Yes |
| 11395 | 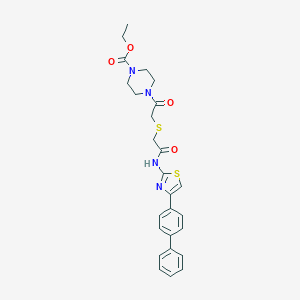 MLS000529745 MLS000529745 | C26H28N4O4S2 | 524.654 | 7 / 1 | 4.0 | No |
| 521806 | 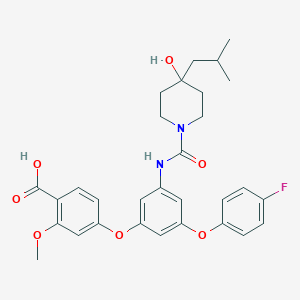 CHEMBL3758974 CHEMBL3758974 | C30H33FN2O7 | 552.599 | 8 / 3 | 5.1 | No |
| 12299 |  MLS000094258 MLS000094258 | C15H10N4O2 | 278.271 | 4 / 0 | 0.5 | Yes |
| 13211 |  CHEMBL1916399 CHEMBL1916399 | C22H15F3N4O5 | 472.38 | 11 / 1 | 3.2 | No |
| 13507 |  CHEMBL224005 CHEMBL224005 | C24H22F3NO3S | 461.499 | 8 / 1 | 2.9 | Yes |
| 13990 |  CHEMBL115131 CHEMBL115131 | C19H33NO3S | 355.537 | 4 / 2 | 2.7 | Yes |
| 14154 | 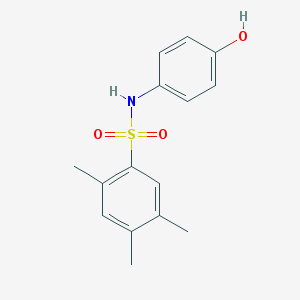 N-(4-hydroxyphenyl)-2,4,5-trimethylbenzenesulfonamide N-(4-hydroxyphenyl)-2,4,5-trimethylbenzenesulfonamide | C15H17NO3S | 291.365 | 4 / 2 | 3.2 | Yes |
| 14401 | 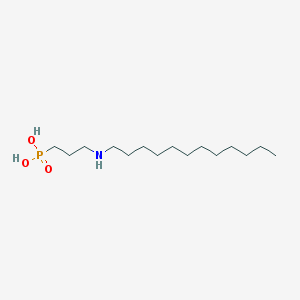 CHEMBL119562 CHEMBL119562 | C15H34NO3P | 307.415 | 4 / 3 | 1.6 | Yes |
| 464672 | 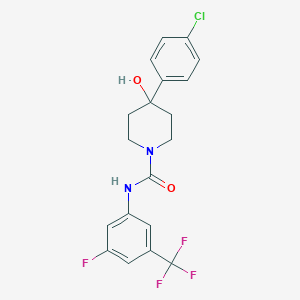 CHEMBL3618192 CHEMBL3618192 | C19H17ClF4N2O2 | 416.801 | 6 / 2 | 3.9 | Yes |
| 15837 | 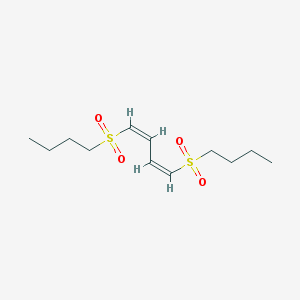 (1Z,3Z)-1,4-bis(butylsulfonyl)buta-1,3-diene (1Z,3Z)-1,4-bis(butylsulfonyl)buta-1,3-diene | C12H22O4S2 | 294.424 | 4 / 0 | 2.1 | Yes |
| 16068 | 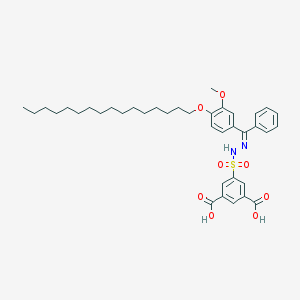 CHEMBL218446 CHEMBL218446 | C38H50N2O8S | 694.884 | 10 / 3 | 11.1 | No |
| 16071 | 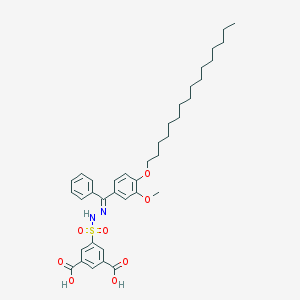 CHEMBL218445 CHEMBL218445 | C38H50N2O8S | 694.884 | 10 / 3 | 11.1 | No |
| 16487 | 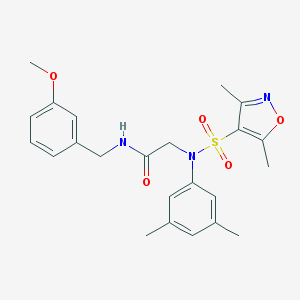 SMR000094367 SMR000094367 | C23H27N3O5S | 457.545 | 7 / 1 | 3.4 | Yes |
| 16493 | 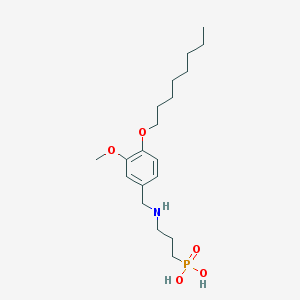 CHEMBL118508 CHEMBL118508 | C19H34NO5P | 387.457 | 6 / 3 | 0.8 | Yes |
| 17993 |  CHEMBL210301 CHEMBL210301 | C20H21N3O4 | 367.405 | 7 / 1 | 3.6 | Yes |
| 18173 |  BAS 09529732 BAS 09529732 | C16H17N3O3 | 299.33 | 4 / 1 | 1.2 | Yes |
| 19541 |  CHEMBL211689 CHEMBL211689 | C21H19F3N2O4 | 420.388 | 9 / 1 | 4.8 | Yes |
| 20104 |  CHEMBL241052 CHEMBL241052 | C21H20N4O4 | 392.415 | 8 / 1 | 3.1 | Yes |
| 442461 |  CHEMBL3359519 CHEMBL3359519 | C24H24F3NO4 | 447.454 | 7 / 1 | 4.8 | Yes |
| 21032 |  CHEMBL324358 CHEMBL324358 | C15H33N2O5P | 352.412 | 6 / 4 | 0.2 | Yes |
| 21322 |  CHEMBL224599 CHEMBL224599 | C23H19BrF3NO3S | 526.368 | 8 / 1 | 3.2 | No |
| 21365 |  CHEMBL332373 CHEMBL332373 | C18H32NO4P | 357.431 | 5 / 3 | 0.9 | Yes |
| 22668 |  CHEMBL1975200 CHEMBL1975200 | C20H30N2O2 | 330.472 | 3 / 0 | 4.9 | Yes |
| 519804 |  CHEMBL3769933 CHEMBL3769933 | C21H24ClN5O3S | 461.965 | 8 / 1 | 1.3 | Yes |
| 442564 |  CHEMBL3401388 CHEMBL3401388 | C29H34FN3O4 | 507.606 | 6 / 2 | 5.7 | No |
| 23277 |  SCHEMBL14338632 SCHEMBL14338632 | C19H34NO4P | 371.458 | 5 / 4 | 1.0 | Yes |
| 23642 | 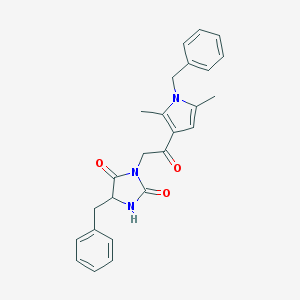 SR-01000759095-1 SR-01000759095-1 | C25H25N3O3 | 415.493 | 3 / 1 | 3.7 | Yes |
| 26760 | 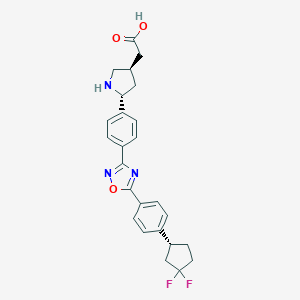 CHEMBL208838 CHEMBL208838 | C25H25F2N3O3 | 453.49 | 8 / 2 | 2.0 | Yes |
| 28713 | 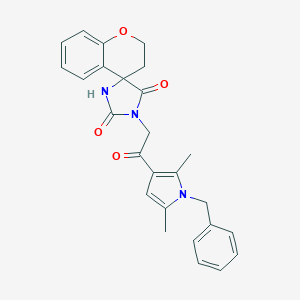 SR-01000759097-1 SR-01000759097-1 | C26H25N3O4 | 443.503 | 4 / 1 | 3.3 | Yes |
| 29344 | 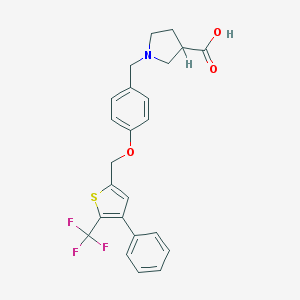 CHEMBL224799 CHEMBL224799 | C24H22F3NO3S | 461.499 | 8 / 1 | 2.9 | Yes |
| 30570 | 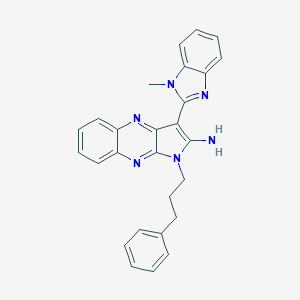 SMR000149318 SMR000149318 | C27H24N6 | 432.531 | 4 / 1 | 4.5 | Yes |
| 32891 | 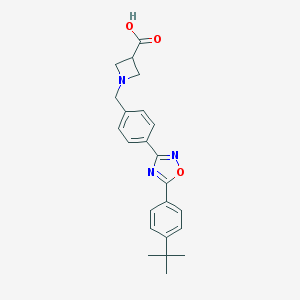 CHEMBL195141 CHEMBL195141 | C23H25N3O3 | 391.471 | 6 / 1 | 2.0 | Yes |
| 33610 | 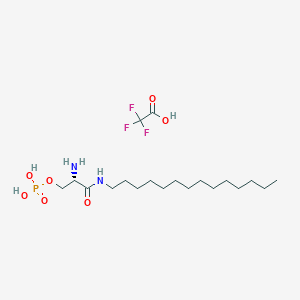 CHEMBL423691 CHEMBL423691 | C19H38F3N2O7P | 494.489 | 11 / 5 | N/A | No |
| 33613 | 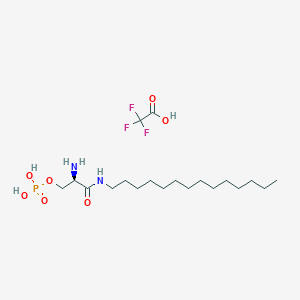 CHEMBL332472 CHEMBL332472 | C19H38F3N2O7P | 494.489 | 11 / 5 | N/A | No |
| 33738 |  SR-01000759123-1 SR-01000759123-1 | C22H22N2O3 | 362.429 | 3 / 1 | 3.3 | Yes |
| 35906 |  CHEMBL119239 CHEMBL119239 | C18H32NO5P | 373.43 | 6 / 4 | 0.6 | Yes |
| 36052 |  SR-01000759093-1 SR-01000759093-1 | C19H21N5OS | 367.471 | 5 / 0 | 3.4 | Yes |
| 467462 |  CHEMBL3703200 CHEMBL3703200 | C36H44FN3O6 | 633.761 | 7 / 3 | 6.1 | No |
| 37382 |  CHEMBL117569 CHEMBL117569 | C21H46NO3P | 391.577 | 4 / 3 | 5.0 | Yes |
| 443168 |  CHEMBL3401377 CHEMBL3401377 | C24H30ClFN2O3 | 448.963 | 4 / 2 | 5.8 | No |
| 37812 |  CHEMBL239659 CHEMBL239659 | C35H43NaO5S2 | 630.834 | 6 / 1 | N/A | No |
| 38426 |  SR-01000759121-1 SR-01000759121-1 | C23H23NO4 | 377.44 | 4 / 1 | 3.7 | Yes |
| 536969 |  SCHEMBL2192987 SCHEMBL2192987 | C24H26N4O5S | 482.555 | 9 / 1 | 3.1 | Yes |
| 536978 |  SCHEMBL1875982 SCHEMBL1875982 | C24H26N4O5S | 482.555 | 9 / 1 | 3.1 | Yes |
| 38697 |  CHEMBL196149 CHEMBL196149 | C23H25N3O3 | 391.471 | 6 / 1 | 1.6 | Yes |
| 39208 |  CHEMBL187692 CHEMBL187692 | C21H44NO2P | 373.562 | 3 / 2 | 4.2 | Yes |
| 41873 | 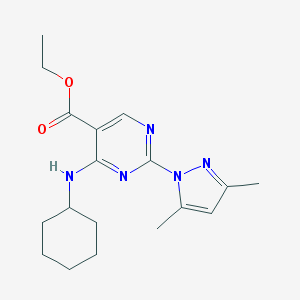 MLS000040640 MLS000040640 | C18H25N5O2 | 343.431 | 6 / 1 | 4.2 | Yes |
| 42399 | 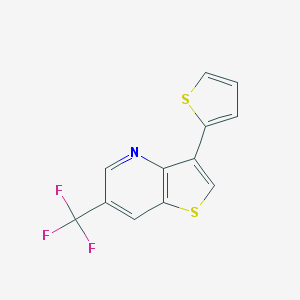 3-(2-thienyl)-6-(trifluoromethyl)thieno[3,2-b]pyridine 3-(2-thienyl)-6-(trifluoromethyl)thieno[3,2-b]pyridine | C12H6F3NS2 | 285.302 | 6 / 0 | 4.2 | Yes |
| 43083 | 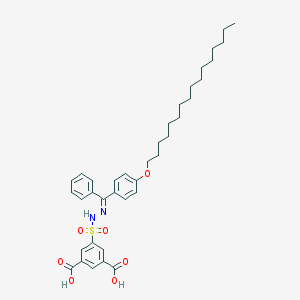 CHEMBL218447 CHEMBL218447 | C37H48N2O7S | 664.858 | 9 / 3 | 11.2 | No |
| 43889 | 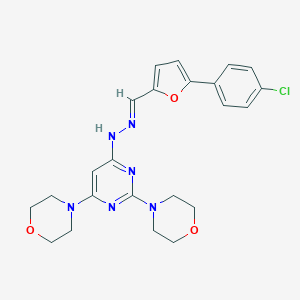 CHEMBL3213728 CHEMBL3213728 | C23H25ClN6O3 | 468.942 | 9 / 1 | 4.0 | Yes |
| 43933 | 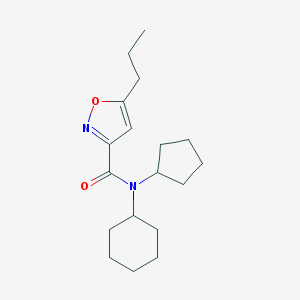 CHEMBL1975908 CHEMBL1975908 | C18H28N2O2 | 304.434 | 3 / 0 | 4.4 | Yes |
| 443412 | 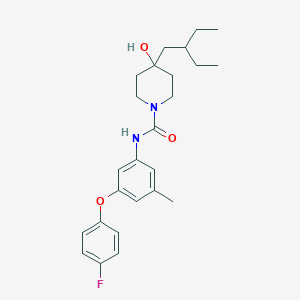 CHEMBL3401376 CHEMBL3401376 | C25H33FN2O3 | 428.548 | 4 / 2 | 5.6 | No |
| 44328 | 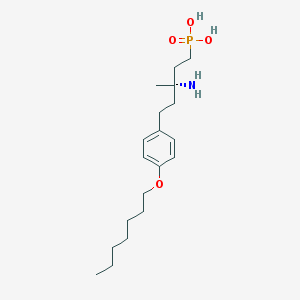 CHEMBL1086170 CHEMBL1086170 | C19H34NO4P | 371.458 | 5 / 3 | 0.9 | Yes |
| 44576 | 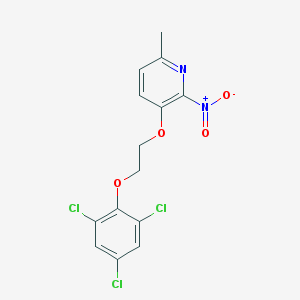 CYM50174 CYM50174 | C14H11Cl3N2O4 | 377.602 | 5 / 0 | 4.9 | Yes |
| 443443 |  CHEMBL3401379 CHEMBL3401379 | C30H34F2N2O4 | 524.609 | 6 / 2 | 6.8 | No |
| 45302 |  CHEMBL1779916 CHEMBL1779916 | C18H17NO2S | 311.399 | 3 / 1 | 4.6 | Yes |
| 45363 |  SR-02000000239 SR-02000000239 | C19H16ClNO2 | 325.792 | 2 / 1 | 5.2 | No |
| 47464 |  CHEMBL91283 CHEMBL91283 | C38H66N2O7P2 | 724.901 | 9 / 7 | N/A | No |
| 47793 | 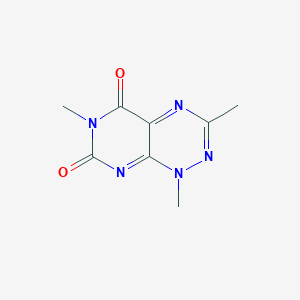 3-Methyltoxoflavin 3-Methyltoxoflavin | C8H9N5O2 | 207.193 | 3 / 0 | -0.7 | Yes |
| 49052 | 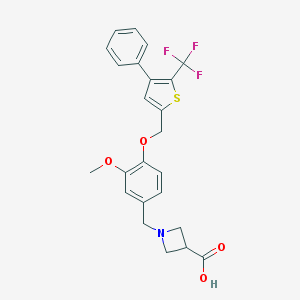 CHEMBL224800 CHEMBL224800 | C24H22F3NO4S | 477.498 | 9 / 1 | 2.5 | Yes |
| 443647 | 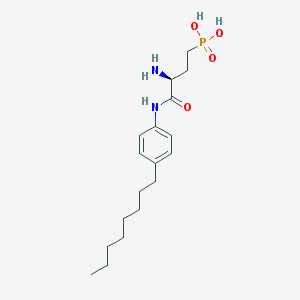 CHEMBL227530 CHEMBL227530 | C18H31N2O4P | 370.43 | 5 / 4 | 0.5 | Yes |
| 443648 | 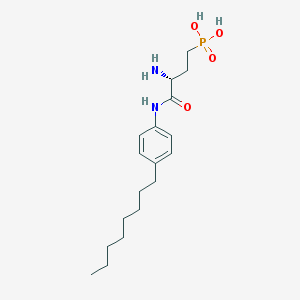 CHEMBL227851 CHEMBL227851 | C18H31N2O4P | 370.43 | 5 / 4 | 0.5 | Yes |
| 50920 |  MLS000083916 MLS000083916 | C24H18N2O6 | 430.416 | 7 / 0 | 3.4 | Yes |
| 51562 |  CHEMBL375488 CHEMBL375488 | C25H24F3NO3S | 475.526 | 8 / 1 | 3.3 | Yes |
| 52217 |  SMR000128117 SMR000128117 | C24H20N2O2 | 368.436 | 2 / 3 | 5.2 | No |
| 469463 |  CHEMBL3618193 CHEMBL3618193 | C19H17F5N2O2 | 400.349 | 7 / 2 | 3.3 | Yes |
| 469607 | 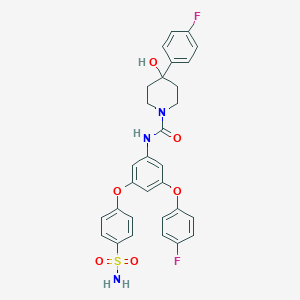 CHEMBL3618200 CHEMBL3618200 | C30H27F2N3O6S | 595.618 | 9 / 3 | 4.1 | No |
| 54725 | 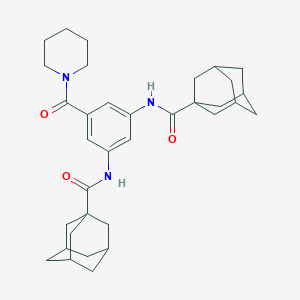 MLS000549502 MLS000549502 | C34H45N3O3 | 543.752 | 3 / 2 | 5.7 | No |
| 469717 | 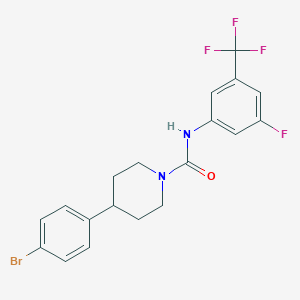 CHEMBL3618195 CHEMBL3618195 | C19H17BrF4N2O | 445.256 | 5 / 1 | 5.0 | Yes |
| 55333 | 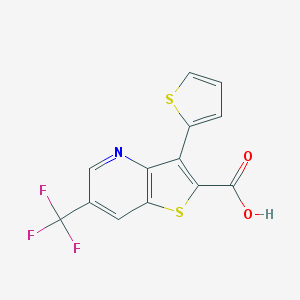 3-(2-thienyl)-6-(trifluoromethyl)thieno[3,2-b]pyridine-2-carboxylic acid 3-(2-thienyl)-6-(trifluoromethyl)thieno[3,2-b]pyridine-2-carboxylic acid | C13H6F3NO2S2 | 329.311 | 8 / 1 | 4.0 | Yes |
| 523154 | 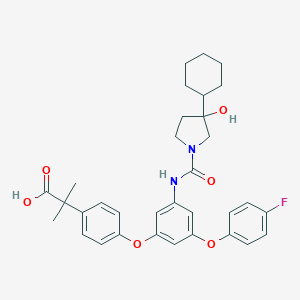 CHEMBL3758803 CHEMBL3758803 | C33H37FN2O6 | 576.665 | 7 / 3 | 6.4 | No |
| 55966 | 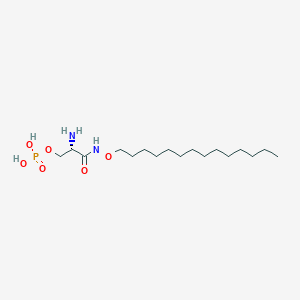 CHEMBL115505 CHEMBL115505 | C17H37N2O6P | 396.465 | 7 / 4 | 1.3 | Yes |
| 56244 | 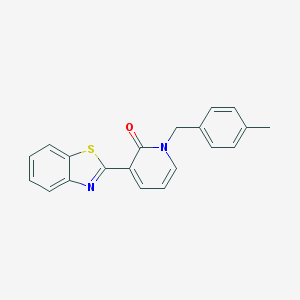 SMR000180629 SMR000180629 | C20H16N2OS | 332.421 | 3 / 0 | 4.4 | Yes |
| 56369 | 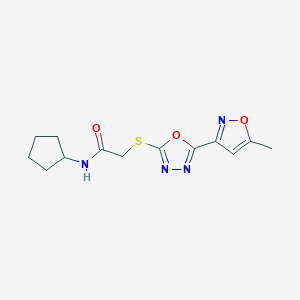 MLS000075167 MLS000075167 | C13H16N4O3S | 308.356 | 7 / 1 | 1.8 | Yes |
| 57885 |  SR-01000759086-1 SR-01000759086-1 | C21H21NO4 | 351.402 | 4 / 0 | 4.1 | Yes |
| 58049 |  methyl 3-(2-pyridinyl)-6-(trifluoromethyl)thieno[3,2-b]pyridine-2-carboxylate methyl 3-(2-pyridinyl)-6-(trifluoromethyl)thieno[3,2-b]pyridine-2-carboxylate | C15H9F3N2O2S | 338.304 | 8 / 0 | 3.6 | Yes |
| 59072 |  CHEMBL1077288 CHEMBL1077288 | C20H33NO3 | 335.488 | 4 / 2 | 2.3 | Yes |
zhanglab![]() zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218