

You can:
| Name | Beta-2 adrenergic receptor |
|---|---|
| Species | Mus musculus (Mouse) |
| Gene | Adrb2 |
| Synonym | beta2-adrenoceptor beta-2 adrenoreceptor Beta-2 adrenoceptor beta-2 adrenergic receptor Adrb-2 [ Show all ] |
| Disease | N/A for non-human GPCRs |
| Length | 418 |
| Amino acid sequence | MGPHGNDSDFLLAPNGSRAPDHDVTQERDEAWVVGMAILMSVIVLAIVFGNVLVITAIAKFERLQTVTNYFIISLACADLVMGLAVVPFGASHILMKMWNFGNFWCEFWTSIDVLCVTASIETLCVIAVDRYVAITSPFKYQSLLTKNKARVVILMVWIVSGLTSFLPIQMHWYRATHKKAIDCYTEETCCDFFTNQAYAIASSIVSFYVPLVVMVFVYSRVFQVAKRQLQKIDKSEGRFHAQNLSQVEQDGRSGHGLRRSSKFCLKEHKALKTLGIIMGTFTLCWLPFFIVNIVHVIRDNLIPKEVYILLNWLGYVNSAFNPLIYCRSPDFRIAFQELLCLRRSSSKTYGNGYSSNSNGRTDYTGEPNTCQLGQEREQELLCEDPPGMEGFVNCQGTVPSLSVDSQGRNCSTNDSPL |
| UniProt | P18762 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | N/A |
| 3D structure model | No available structures or models |
| BioLiP | N/A |
| Therapeutic Target Database | N/A |
| ChEMBL | CHEMBL3707 |
| IUPHAR | N/A |
| DrugBank | N/A |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 459289 | 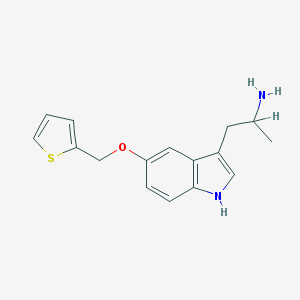 BW-723C86 BW-723C86 | C16H18N2OS | 286.393 | 3 / 2 | 3.0 | Yes |
| 69360 | 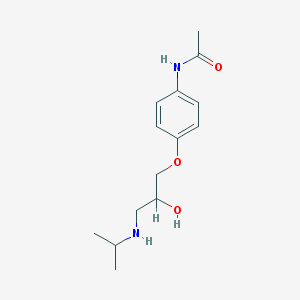 practolol practolol | C14H22N2O3 | 266.341 | 4 / 3 | 0.8 | Yes |
| 108256 | 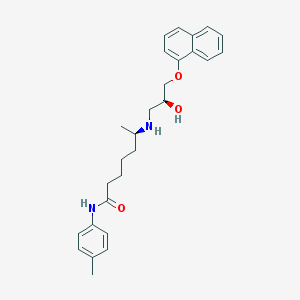 CHEMBL70679 CHEMBL70679 | C27H34N2O3 | 434.58 | 4 / 3 | 4.9 | Yes |
| 108257 | 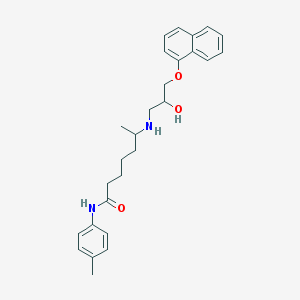 CHEMBL304961 CHEMBL304961 | C27H34N2O3 | 434.58 | 4 / 3 | 4.9 | Yes |
| 108258 | 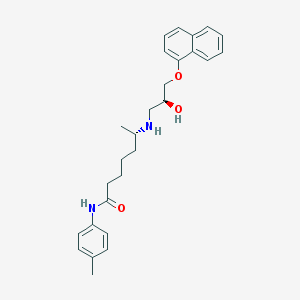 CHEMBL71765 CHEMBL71765 | C27H34N2O3 | 434.58 | 4 / 3 | 4.9 | Yes |
| 108259 | 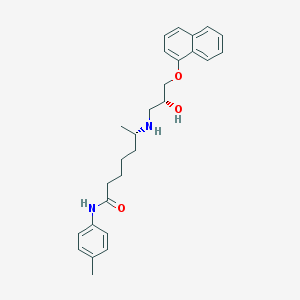 CHEMBL308698 CHEMBL308698 | C27H34N2O3 | 434.58 | 4 / 3 | 4.9 | Yes |
| 108260 | 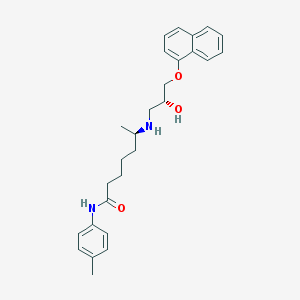 CHEMBL72113 CHEMBL72113 | C27H34N2O3 | 434.58 | 4 / 3 | 4.9 | Yes |
| 460456 | 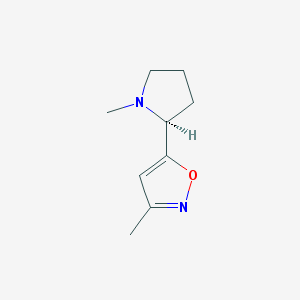 Abt-418 Abt-418 | C9H14N2O | 166.224 | 3 / 0 | 1.2 | Yes |
| 205431 | 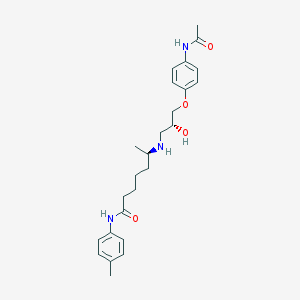 CHEMBL74122 CHEMBL74122 | C25H35N3O4 | 441.572 | 5 / 4 | 2.7 | Yes |
| 205432 | 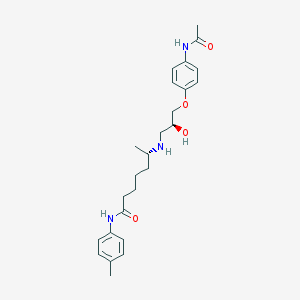 CHEMBL71734 CHEMBL71734 | C25H35N3O4 | 441.572 | 5 / 4 | 2.7 | Yes |
| 205433 | 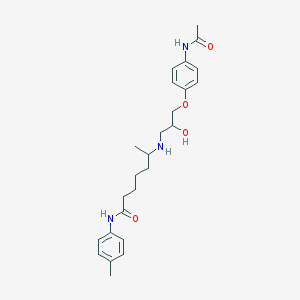 CHEMBL70809 CHEMBL70809 | C25H35N3O4 | 441.572 | 5 / 4 | 2.7 | Yes |
| 205434 | 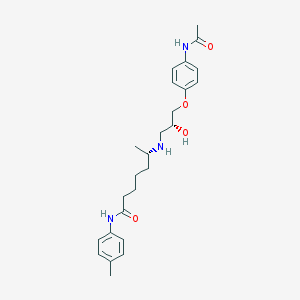 CHEMBL69449 CHEMBL69449 | C25H35N3O4 | 441.572 | 5 / 4 | 2.7 | Yes |
| 205435 | 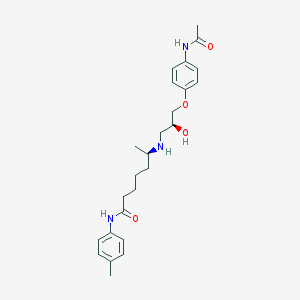 CHEMBL305051 CHEMBL305051 | C25H35N3O4 | 441.572 | 5 / 4 | 2.7 | Yes |
| 218064 | 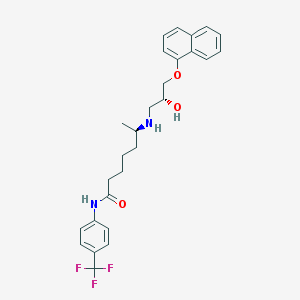 CHEMBL71586 CHEMBL71586 | C27H31F3N2O3 | 488.551 | 7 / 3 | 5.4 | No |
| 218065 | 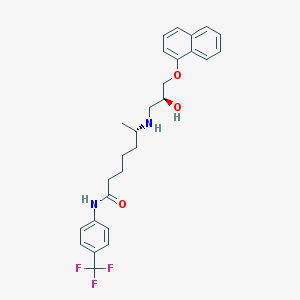 CHEMBL72707 CHEMBL72707 | C27H31F3N2O3 | 488.551 | 7 / 3 | 5.4 | No |
| 218066 | 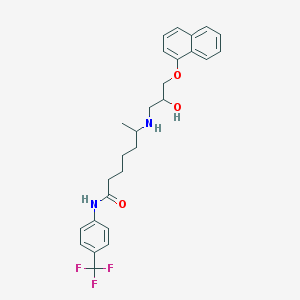 CHEMBL70840 CHEMBL70840 | C27H31F3N2O3 | 488.551 | 7 / 3 | 5.4 | No |
| 218067 | 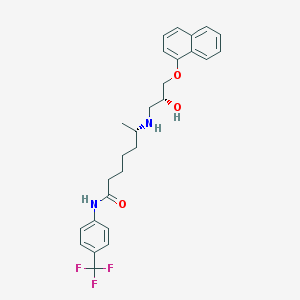 CHEMBL73722 CHEMBL73722 | C27H31F3N2O3 | 488.551 | 7 / 3 | 5.4 | No |
| 218068 | 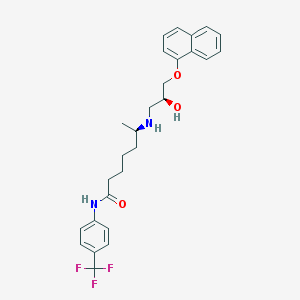 CHEMBL70539 CHEMBL70539 | C27H31F3N2O3 | 488.551 | 7 / 3 | 5.4 | No |
| 377516 | 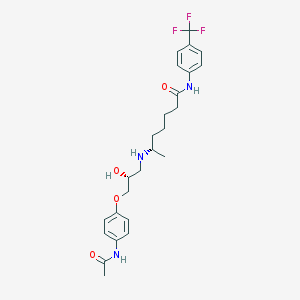 CHEMBL69036 CHEMBL69036 | C25H32F3N3O4 | 495.543 | 8 / 4 | 3.2 | Yes |
| 377517 | 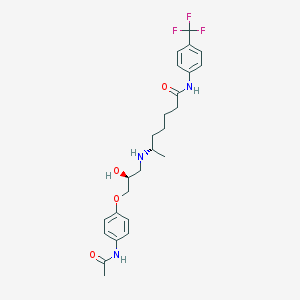 CHEMBL71432 CHEMBL71432 | C25H32F3N3O4 | 495.543 | 8 / 4 | 3.2 | Yes |
| 377518 | 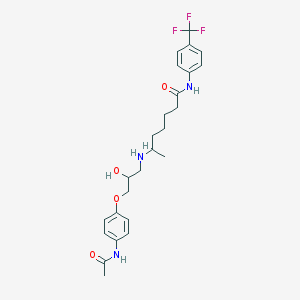 CHEMBL70217 CHEMBL70217 | C25H32F3N3O4 | 495.543 | 8 / 4 | 3.2 | Yes |
| 377519 | 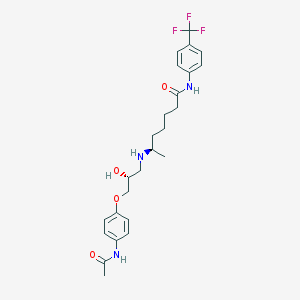 CHEMBL421682 CHEMBL421682 | C25H32F3N3O4 | 495.543 | 8 / 4 | 3.2 | Yes |
| 377520 | 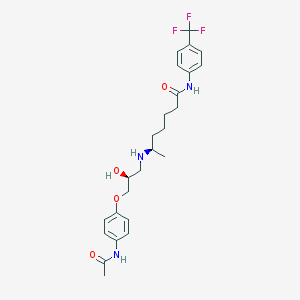 CHEMBL306237 CHEMBL306237 | C25H32F3N3O4 | 495.543 | 8 / 4 | 3.2 | Yes |
| 462581 | 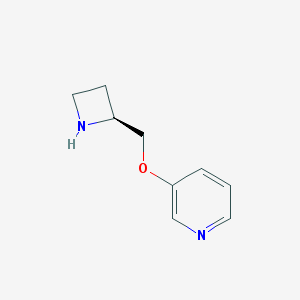 A-85380 A-85380 | C9H12N2O | 164.208 | 3 / 1 | 0.6 | Yes |
zhanglab![]() zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218