

You can:
| Name | Gonadotropin-releasing hormone receptor |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | GNRHR |
| Synonym | LHRHR GnRH I receptor Type I GnRHR gnRH receptor GnRH-R [ Show all ] |
| Disease | Uterine leiomyoma Hypothalamic hypogonadism Ovulation Prostate cancer Prostate disease [ Show all ] |
| Length | 328 |
| Amino acid sequence | MANSASPEQNQNHCSAINNSIPLMQGNLPTLTLSGKIRVTVTFFLFLLSATFNASFLLKLQKWTQKKEKGKKLSRMKLLLKHLTLANLLETLIVMPLDGMWNITVQWYAGELLCKVLSYLKLFSMYAPAFMMVVISLDRSLAITRPLALKSNSKVGQSMVGLAWILSSVFAGPQLYIFRMIHLADSSGQTKVFSQCVTHCSFSQWWHQAFYNFFTFSCLFIIPLFIMLICNAKIIFTLTRVLHQDPHELQLNQSKNNIPRARLKTLKMTVAFATSFTVCWTPYYVLGIWYWFDPEMLNRLSDPVNHFFFLFAFLNPCFDPLIYGYFSL |
| UniProt | P30968 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | P30968 |
| 3D structure model | This predicted structure model is from GPCR-EXP P30968. |
| BioLiP | N/A |
| Therapeutic Target Database | T12475 |
| ChEMBL | CHEMBL1855 |
| IUPHAR | 256 |
| DrugBank | BE0000203 |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 15 | 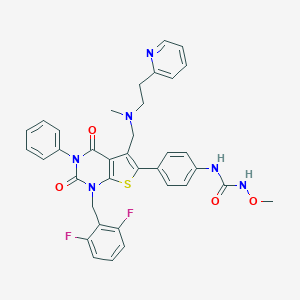 CHEMBL1800663 CHEMBL1800663 | C36H32F2N6O4S | 682.747 | 9 / 2 | 5.5 | No |
| 919 | 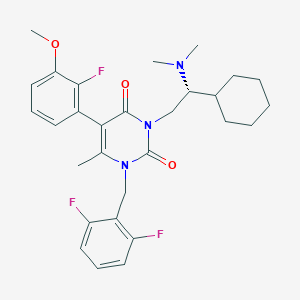 CHEMBL119774 CHEMBL119774 | C29H34F3N3O3 | 529.604 | 7 / 0 | 5.6 | No |
| 1110 | 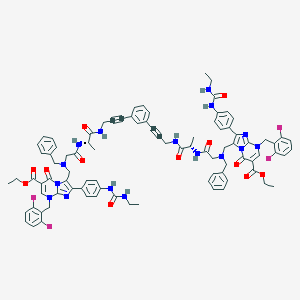 CHEMBL437260 CHEMBL437260 | C88H86F4N16O12 | 1635.75 | 22 / 8 | 9.5 | No |
| 1572 | 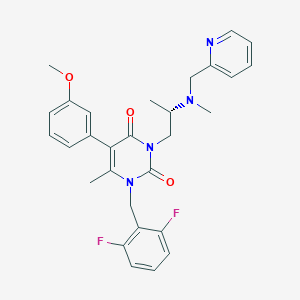 CHEMBL324759 CHEMBL324759 | C29H30F2N4O3 | 520.581 | 7 / 0 | 4.0 | No |
| 1574 | 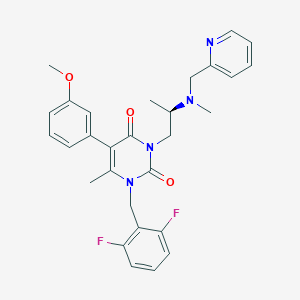 CHEMBL112466 CHEMBL112466 | C29H30F2N4O3 | 520.581 | 7 / 0 | 4.0 | No |
| 1575 | 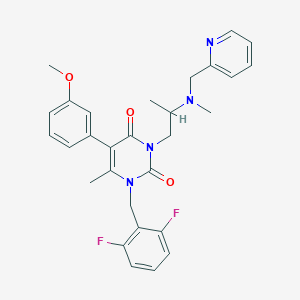 CHEMBL113151 CHEMBL113151 | C29H30F2N4O3 | 520.581 | 7 / 0 | 4.0 | No |
| 2247 | 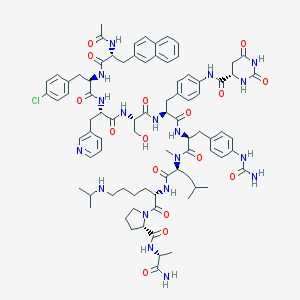 CHEMBL265879 CHEMBL265879 | C83H105ClN18O16 | 1646.31 | 18 / 16 | 3.7 | No |
| 2357 | 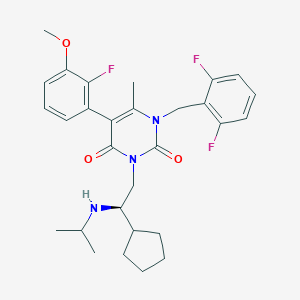 CHEMBL117504 CHEMBL117504 | C29H34F3N3O3 | 529.604 | 7 / 1 | 5.4 | No |
| 2998 | 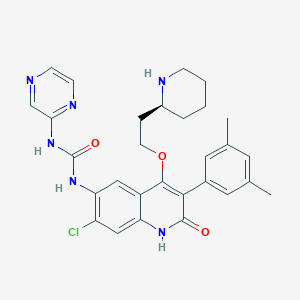 CHEMBL416651 CHEMBL416651 | C29H31ClN6O3 | 547.056 | 6 / 4 | 3.6 | No |
| 3246 | 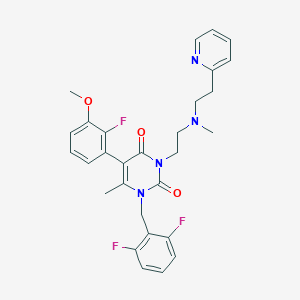 CHEMBL417645 CHEMBL417645 | C29H29F3N4O3 | 538.571 | 8 / 0 | 4.1 | No |
| 3366 | 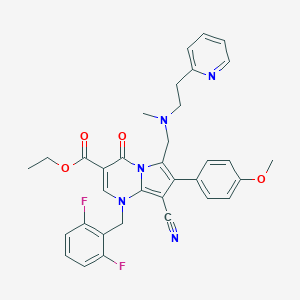 CHEMBL130607 CHEMBL130607 | C34H31F2N5O4 | 611.65 | 10 / 0 | 5.1 | No |
| 3635 | 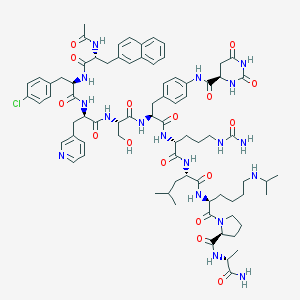 CHEMBL219461 CHEMBL219461 | C78H103ClN18O16 | 1584.24 | 18 / 17 | 2.4 | No |
| 3671 | 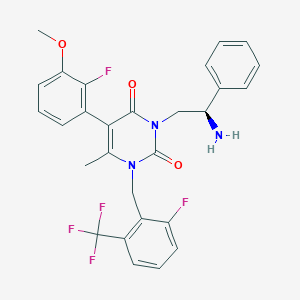 CHEMBL468551 CHEMBL468551 | C28H24F5N3O3 | 545.51 | 9 / 1 | 4.5 | No |
| 3990 | 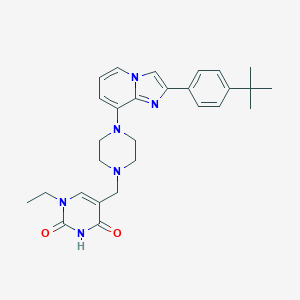 CHEMBL1091579 CHEMBL1091579 | C28H34N6O2 | 486.62 | 5 / 1 | 4.2 | Yes |
| 4039 | 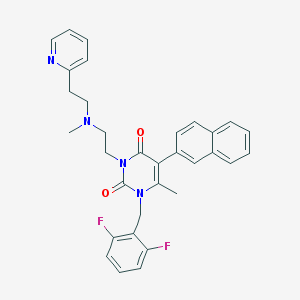 CHEMBL22937 CHEMBL22937 | C32H30F2N4O2 | 540.615 | 6 / 0 | 5.3 | No |
| 4274 | 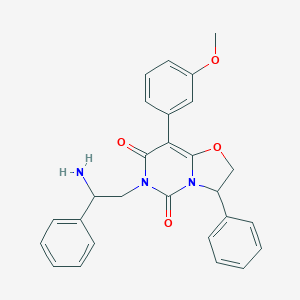 CHEMBL181582 CHEMBL181582 | C27H25N3O4 | 455.514 | 5 / 1 | 3.1 | Yes |
| 4621 | 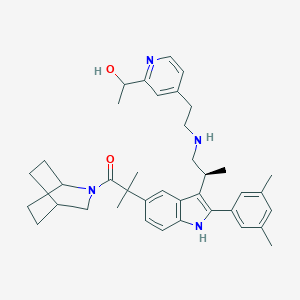 CHEMBL113432 CHEMBL113432 | C39H50N4O2 | 606.855 | 4 / 3 | 6.7 | No |
| 4743 | 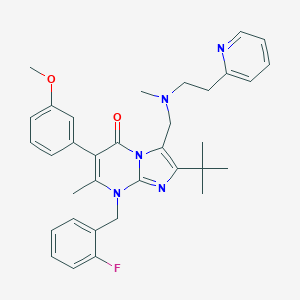 CHEMBL50546 CHEMBL50546 | C34H38FN5O2 | 567.709 | 7 / 0 | 6.3 | No |
| 5350 | 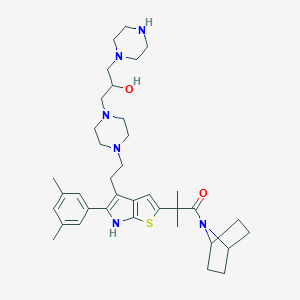 CHEMBL401418 CHEMBL401418 | C37H54N6O2S | 646.939 | 7 / 3 | 4.8 | No |
| 6603 | 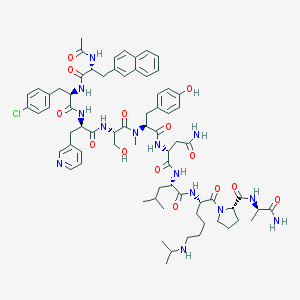 Abarelix Abarelix | C72H95ClN14O14 | 1416.09 | 16 / 13 | 3.7 | No |
| 7096 | 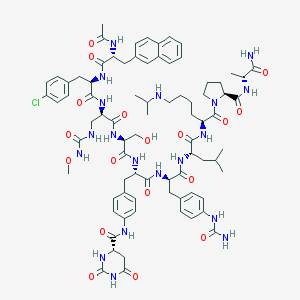 CHEMBL411381 CHEMBL411381 | C79H104ClN19O18 | 1643.27 | 19 / 19 | 1.8 | No |
| 7195 | 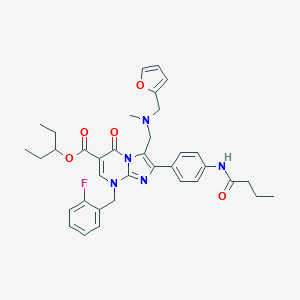 CHEMBL71793 CHEMBL71793 | C36H40FN5O5 | 641.744 | 9 / 1 | 5.9 | No |
| 7425 | 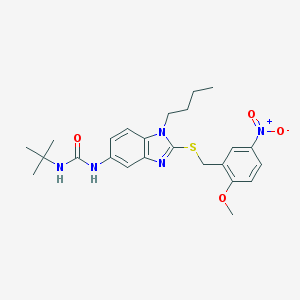 CHEMBL372316 CHEMBL372316 | C24H31N5O4S | 485.603 | 6 / 2 | 4.8 | Yes |
| 7636 | 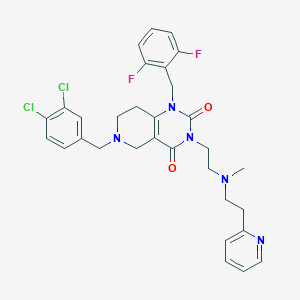 CHEMBL392604 CHEMBL392604 | C31H31Cl2F2N5O2 | 614.519 | 7 / 0 | 4.7 | No |
| 8740 | 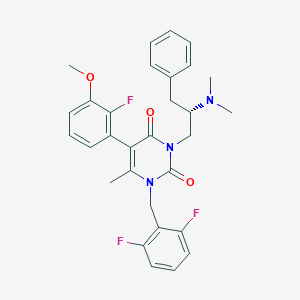 CHEMBL116074 CHEMBL116074 | C30H30F3N3O3 | 537.583 | 7 / 0 | 5.2 | No |
| 9009 | 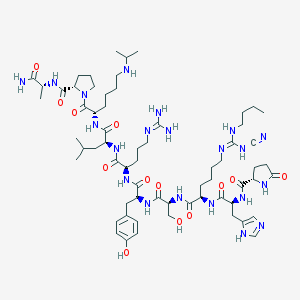 CHEMBL407023 CHEMBL407023 | C64H103N21O13 | 1374.66 | 18 / 18 | -1.0 | No |
| 9930 | 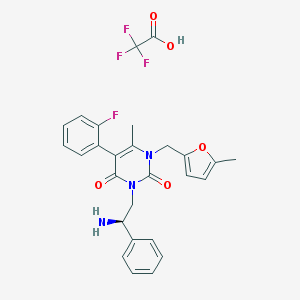 CHEMBL73454 CHEMBL73454 | C27H25F4N3O5 | 547.507 | 10 / 2 | N/A | No |
| 10377 | 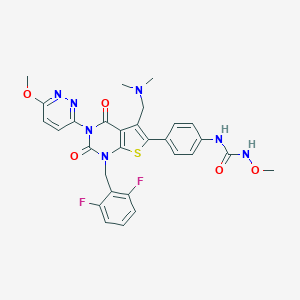 Relugolix Relugolix | C29H27F2N7O5S | 623.636 | 11 / 2 | 3.2 | No |
| 10571 | 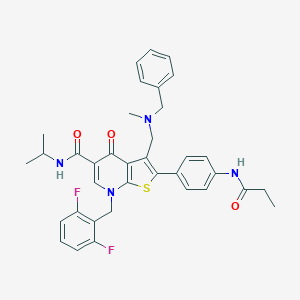 CHEMBL211731 CHEMBL211731 | C36H36F2N4O3S | 642.766 | 8 / 2 | 6.9 | No |
| 10617 | 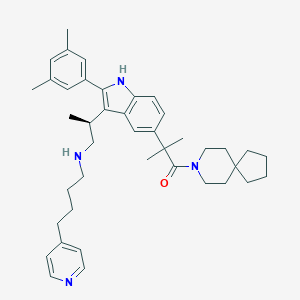 CHEMBL420051 CHEMBL420051 | C41H54N4O | 618.91 | 3 / 2 | 9.1 | No |
| 11236 | 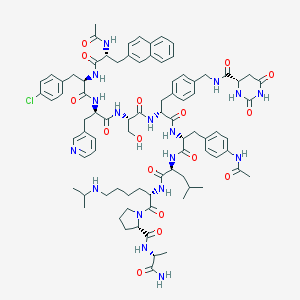 CHEMBL411565 CHEMBL411565 | C84H106ClN17O16 | 1645.32 | 18 / 16 | 4.0 | No |
| 11297 | 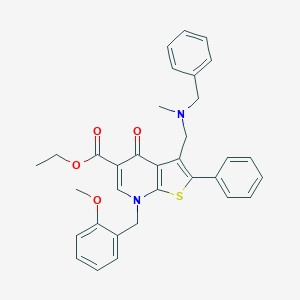 CHEMBL212207 CHEMBL212207 | C33H32N2O4S | 552.689 | 7 / 0 | 6.6 | No |
| 11302 | 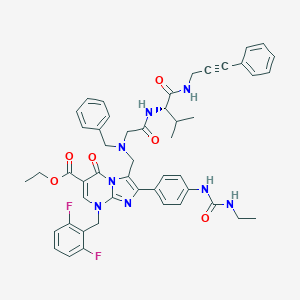 CHEMBL439076 CHEMBL439076 | C49H50F2N8O6 | 884.986 | 11 / 4 | 6.7 | No |
| 11457 | 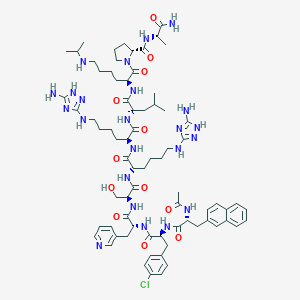 CHEMBL2370818 CHEMBL2370818 | C74H106ClN23O12 | 1545.26 | 22 / 18 | 4.7 | No |
| 11588 | 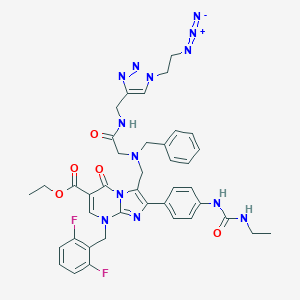 CHEMBL235180 CHEMBL235180 | C40H41F2N13O5 | 821.851 | 14 / 3 | 4.2 | No |
| 12009 | 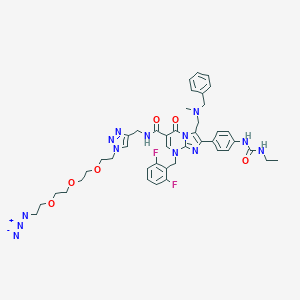 CHEMBL238377 CHEMBL238377 | C43H49F2N13O6 | 881.947 | 15 / 3 | 4.1 | No |
| 12049 | 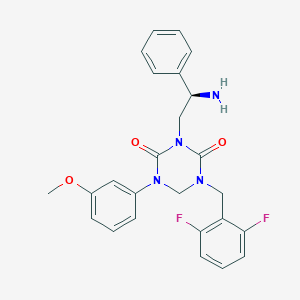 CHEMBL2113012 CHEMBL2113012 | C25H24F2N4O3 | 466.489 | 6 / 1 | 3.2 | Yes |
| 12143 | 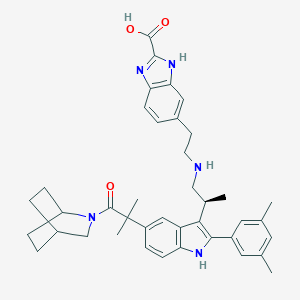 CHEMBL352051 CHEMBL352051 | C40H47N5O3 | 645.848 | 5 / 4 | 5.3 | No |
| 12236 | 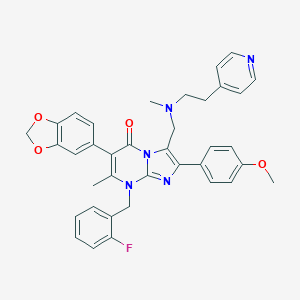 CHEMBL68985 CHEMBL68985 | C37H34FN5O4 | 631.708 | 9 / 0 | 6.0 | No |
| 12379 | 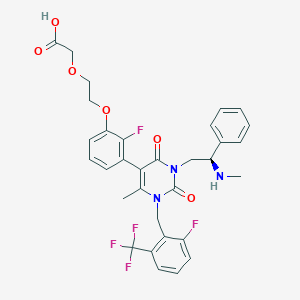 CHEMBL506338 CHEMBL506338 | C32H30F5N3O6 | 647.599 | 12 / 2 | 2.2 | No |
| 12509 | 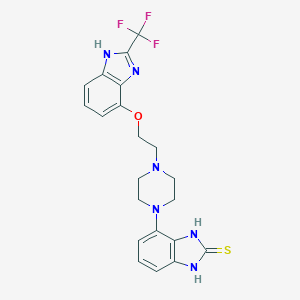 CHEMBL454088 CHEMBL454088 | C21H21F3N6OS | 462.495 | 8 / 3 | 3.2 | Yes |
| 12557 | 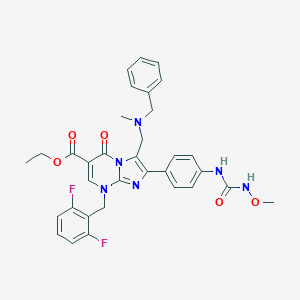 CHEMBL291896 CHEMBL291896 | C33H32F2N6O5 | 630.653 | 10 / 2 | 4.4 | No |
| 12669 | 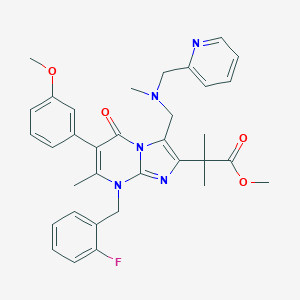 CHEMBL299348 CHEMBL299348 | C34H36FN5O4 | 597.691 | 9 / 0 | 4.9 | No |
| 13576 | 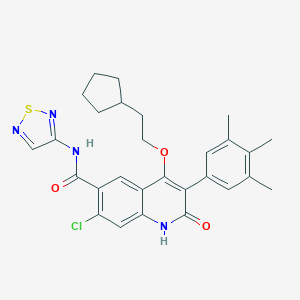 CHEMBL185438 CHEMBL185438 | C28H29ClN4O3S | 537.075 | 6 / 2 | 6.5 | No |
| 13588 | 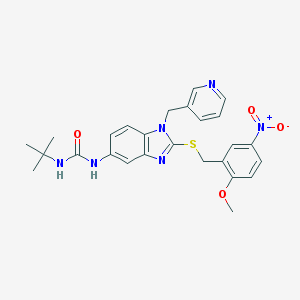 CHEMBL364933 CHEMBL364933 | C26H28N6O4S | 520.608 | 7 / 2 | 4.1 | No |
| 13796 | 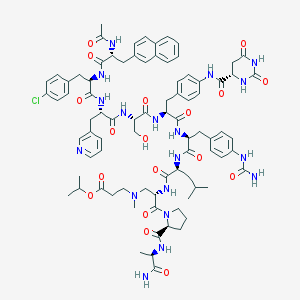 CHEMBL265020 CHEMBL265020 | C83H103ClN18O18 | 1676.3 | 20 / 16 | 2.7 | No |
| 14422 | 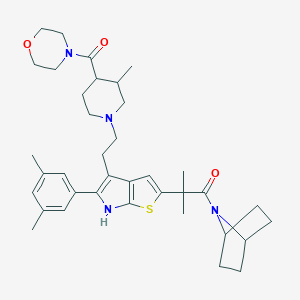 CHEMBL399857 CHEMBL399857 | C37H50N4O3S | 630.892 | 5 / 1 | 6.2 | No |
| 14757 | 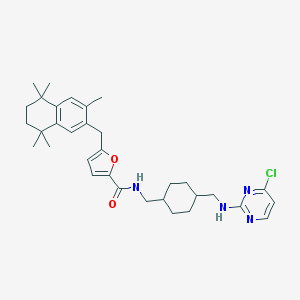 CHEMBL120834 CHEMBL120834 | C33H43ClN4O2 | 563.183 | 5 / 2 | 9.0 | No |
| 14986 | 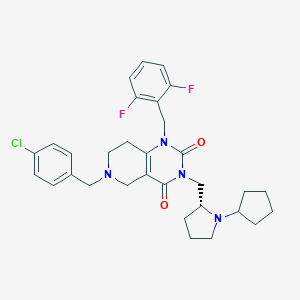 CHEMBL235519 CHEMBL235519 | C31H35ClF2N4O2 | 569.094 | 6 / 0 | 5.0 | No |
| 15030 | 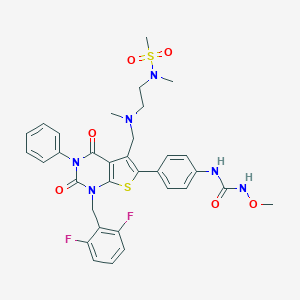 CHEMBL1800667 CHEMBL1800667 | C33H34F2N6O6S2 | 712.788 | 11 / 2 | 3.9 | No |
| 15218 | 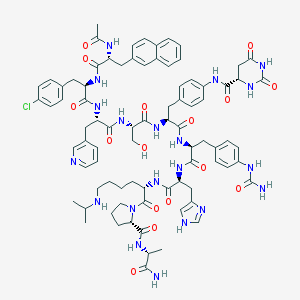 CID 44398121 CID 44398121 | C82H99ClN20O16 | 1656.27 | 19 / 18 | 1.9 | No |
| 15233 | 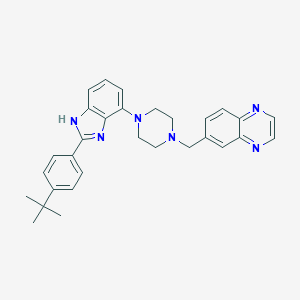 WAY-207024 WAY-207024 | C30H32N6 | 476.628 | 5 / 1 | 5.5 | No |
| 15618 | 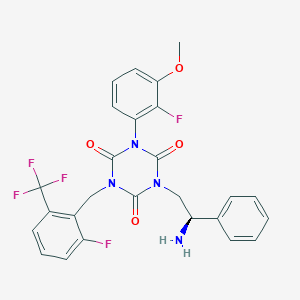 CHEMBL175711 CHEMBL175711 | C26H21F5N4O4 | 548.47 | 10 / 1 | 4.1 | No |
| 15720 | 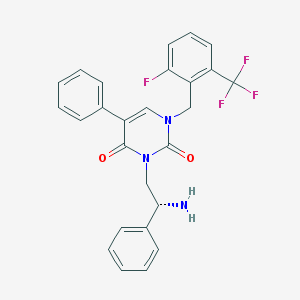 CHEMBL365985 CHEMBL365985 | C26H21F4N3O2 | 483.467 | 7 / 1 | 4.1 | Yes |
| 15928 | 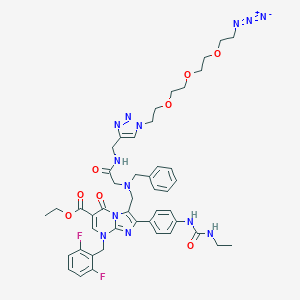 CHEMBL236221 CHEMBL236221 | C46H53F2N13O8 | 954.01 | 17 / 3 | 3.8 | No |
| 16615 | 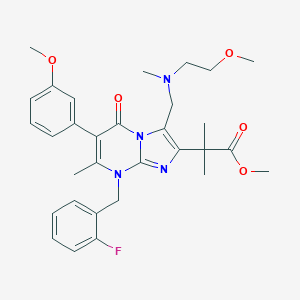 CHEMBL301103 CHEMBL301103 | C31H37FN4O5 | 564.658 | 9 / 0 | 4.3 | No |
| 16672 | 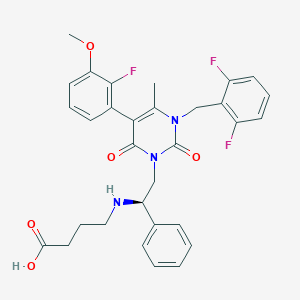 CHEMBL506877 CHEMBL506877 | C31H30F3N3O5 | 581.592 | 9 / 2 | 1.8 | No |
| 17182 | 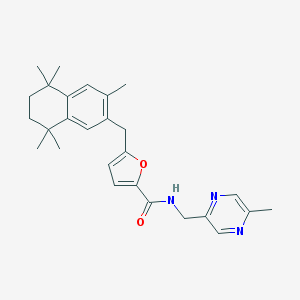 CHEMBL116246 CHEMBL116246 | C27H33N3O2 | 431.58 | 4 / 1 | 5.8 | No |
| 17656 | 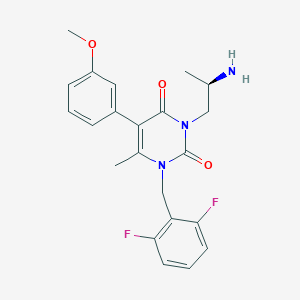 CHEMBL326510 CHEMBL326510 | C22H23F2N3O3 | 415.441 | 6 / 1 | 2.6 | Yes |
| 17657 | 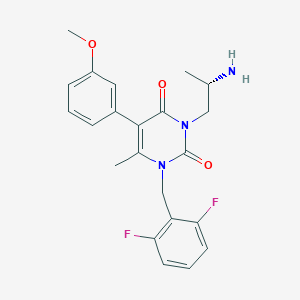 CHEMBL322283 CHEMBL322283 | C22H23F2N3O3 | 415.441 | 6 / 1 | 2.6 | Yes |
| 17743 | 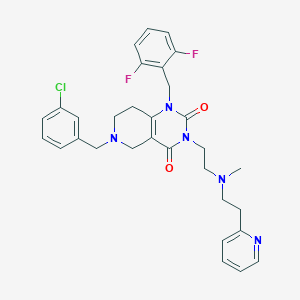 CHEMBL236331 CHEMBL236331 | C31H32ClF2N5O2 | 580.077 | 7 / 0 | 4.1 | No |
| 17752 | 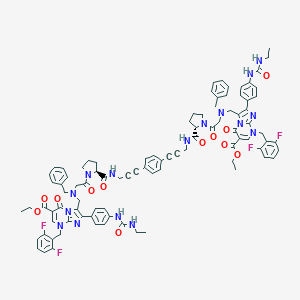 CHEMBL260750 CHEMBL260750 | C92H90F4N16O12 | 1687.83 | 22 / 6 | 10.1 | No |
| 17850 | 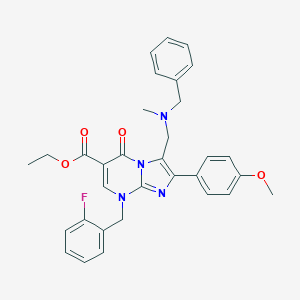 CHEMBL68664 CHEMBL68664 | C32H31FN4O4 | 554.622 | 8 / 0 | 5.2 | No |
| 18753 | 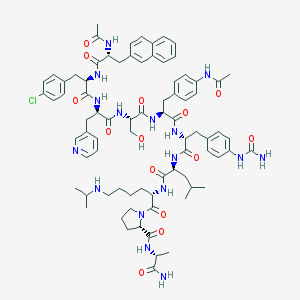 CHEMBL406337 CHEMBL406337 | C79H101ClN16O14 | 1534.22 | 16 / 15 | 4.8 | No |
| 18859 | 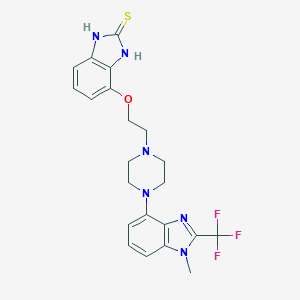 CHEMBL452541 CHEMBL452541 | C22H23F3N6OS | 476.522 | 8 / 2 | 3.2 | Yes |
| 19092 | 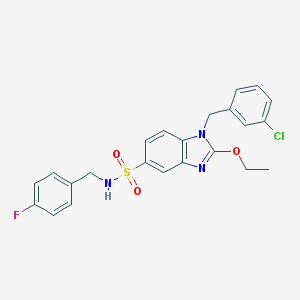 CHEMBL181864 CHEMBL181864 | C23H21ClFN3O3S | 473.947 | 6 / 1 | 4.9 | Yes |
| 19880 | 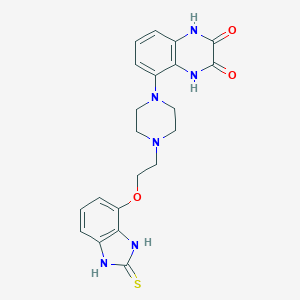 CHEMBL454089 CHEMBL454089 | C21H22N6O3S | 438.506 | 6 / 4 | 1.3 | Yes |
| 20308 | 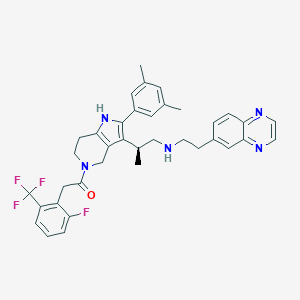 CHEMBL395225 CHEMBL395225 | C37H37F4N5O | 643.731 | 8 / 2 | 6.4 | No |
| 20572 | 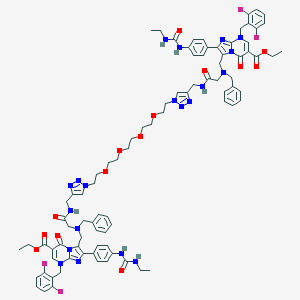 CHEMBL392008 CHEMBL392008 | C86H94F4N20O14 | 1707.82 | 28 / 6 | 5.8 | No |
| 20759 | 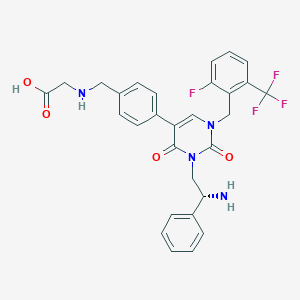 CHEMBL499849 CHEMBL499849 | C29H26F4N4O4 | 570.545 | 10 / 3 | 0.8 | No |
| 21457 | 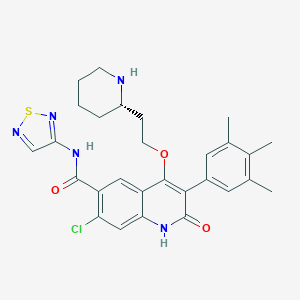 CHEMBL433594 CHEMBL433594 | C28H30ClN5O3S | 552.09 | 7 / 3 | 4.8 | No |
| 21723 | 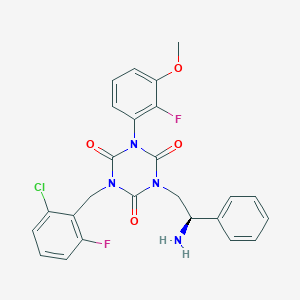 CHEMBL180372 CHEMBL180372 | C25H21ClF2N4O4 | 514.914 | 7 / 1 | 3.8 | No |
| 21811 | 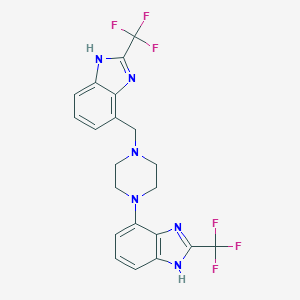 CHEMBL487627 CHEMBL487627 | C21H18F6N6 | 468.407 | 10 / 2 | 4.3 | Yes |
| 21878 | 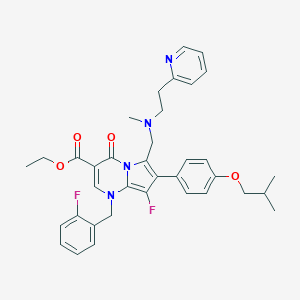 CHEMBL117069 CHEMBL117069 | C36H38F2N4O4 | 628.721 | 9 / 0 | 6.7 | No |
| 22588 | 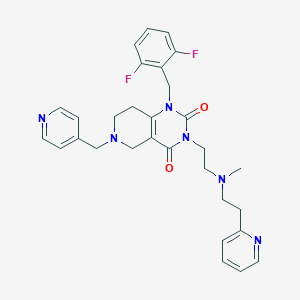 CHEMBL235302 CHEMBL235302 | C30H32F2N6O2 | 546.623 | 8 / 0 | 2.4 | No |
| 23443 | 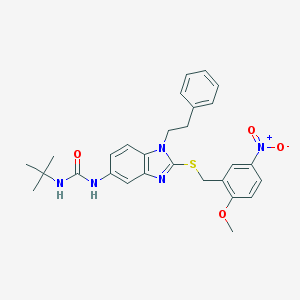 CHEMBL364246 CHEMBL364246 | C28H31N5O4S | 533.647 | 6 / 2 | 5.5 | No |
| 24221 | 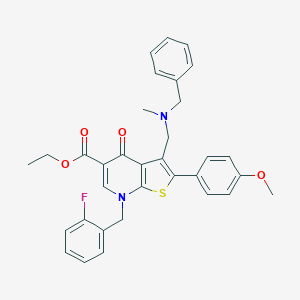 CHEMBL210277 CHEMBL210277 | C33H31FN2O4S | 570.679 | 8 / 0 | 6.7 | No |
| 24302 | 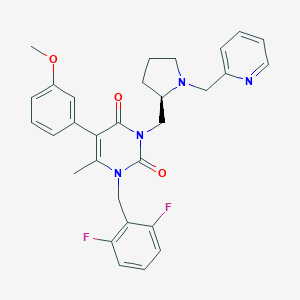 CHEMBL325326 CHEMBL325326 | C30H30F2N4O3 | 532.592 | 7 / 0 | 4.1 | No |
| 24303 | 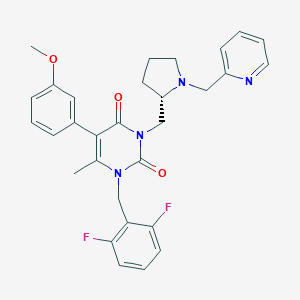 CHEMBL431248 CHEMBL431248 | C30H30F2N4O3 | 532.592 | 7 / 0 | 4.1 | No |
| 25166 | 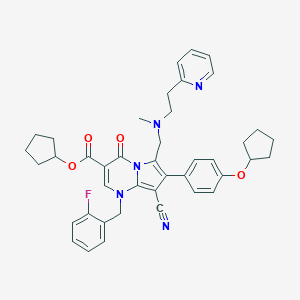 CHEMBL131665 CHEMBL131665 | C41H42FN5O4 | 687.816 | 9 / 0 | 7.2 | No |
| 548169 | 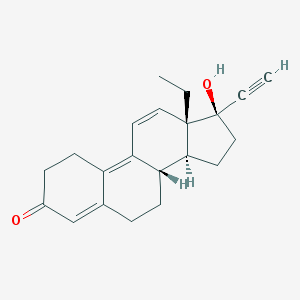 Gestrinone Gestrinone | C21H24O2 | 308.421 | 2 / 1 | 2.2 | Yes |
| 25366 | 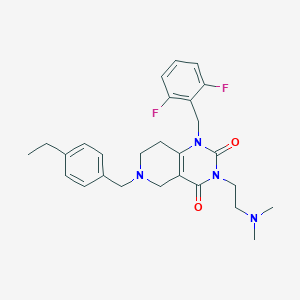 CHEMBL392688 CHEMBL392688 | C27H32F2N4O2 | 482.576 | 6 / 0 | 3.3 | Yes |
| 25416 | 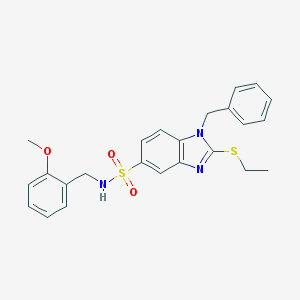 CHEMBL468348 CHEMBL468348 | C24H25N3O3S2 | 467.602 | 6 / 1 | 4.7 | Yes |
| 25485 | 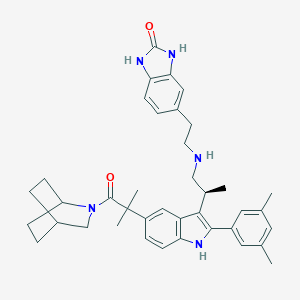 CHEMBL165456 CHEMBL165456 | C39H47N5O2 | 617.838 | 3 / 4 | 6.8 | No |
| 25549 | 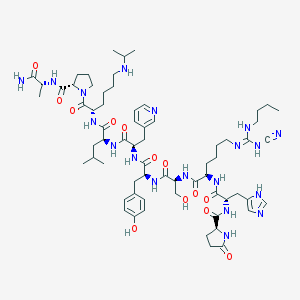 CHEMBL267862 CHEMBL267862 | C66H99N19O13 | 1366.64 | 18 / 16 | 0.7 | No |
| 25599 | 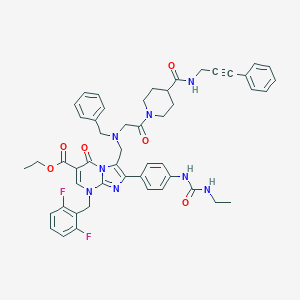 CHEMBL415279 CHEMBL415279 | C50H50F2N8O6 | 896.997 | 11 / 3 | 5.9 | No |
| 548172 | 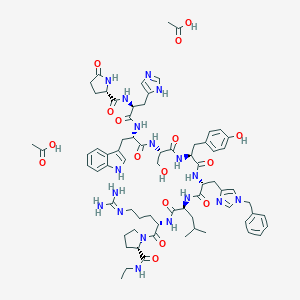 Vantas Vantas | C70H94N18O16 | 1443.63 | 19 / 17 | N/A | No |
| 26130 | 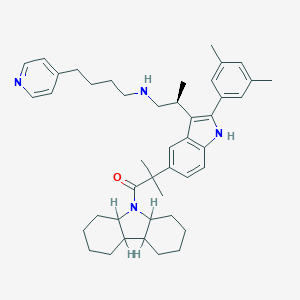 CHEMBL313004 CHEMBL313004 | C44H58N4O | 658.975 | 3 / 2 | 9.7 | No |
| 26403 | 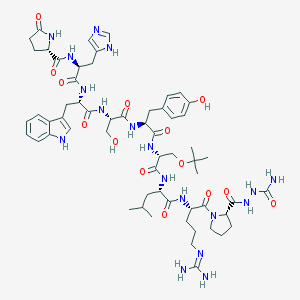 goserelin goserelin | C59H84N18O14 | 1269.43 | 16 / 17 | -1.5 | No |
| 26465 | 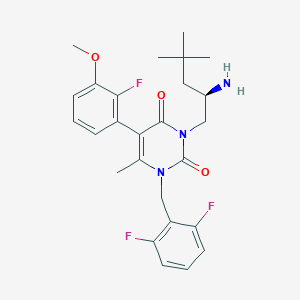 CHEMBL115965 CHEMBL115965 | C26H30F3N3O3 | 489.539 | 7 / 1 | 4.4 | Yes |
| 26797 | 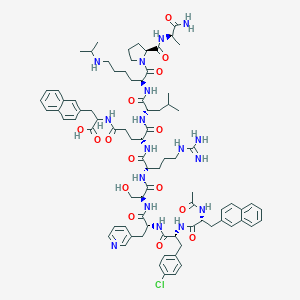 CHEMBL384898 CHEMBL384898 | C82H108ClN17O15 | 1607.32 | 18 / 17 | 2.8 | No |
| 26798 | 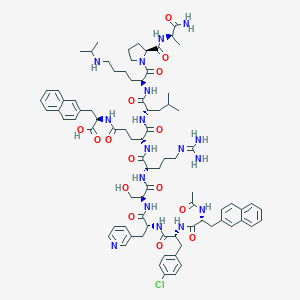 CHEMBL2370890 CHEMBL2370890 | C82H108ClN17O15 | 1607.32 | 18 / 16 | 2.4 | No |
| 459454 | 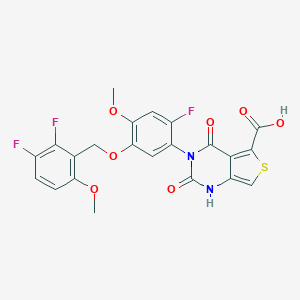 UNII-7CDW97HUEX UNII-7CDW97HUEX | C22H15F3N2O7S | 508.424 | 11 / 2 | 3.4 | No |
| 27547 | 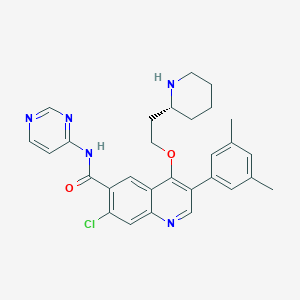 CHEMBL1766108 CHEMBL1766108 | C29H30ClN5O2 | 516.042 | 6 / 2 | 5.4 | No |
| 27867 | 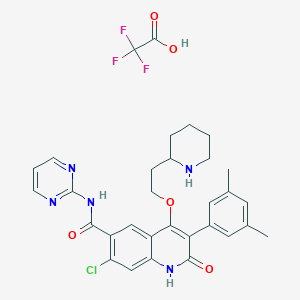 CHEMBL342396 CHEMBL342396 | C31H31ClF3N5O5 | 646.064 | 11 / 4 | N/A | No |
| 28328 | 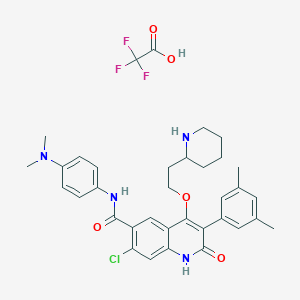 CHEMBL342908 CHEMBL342908 | C35H38ClF3N4O5 | 687.157 | 10 / 4 | N/A | No |
| 28481 | 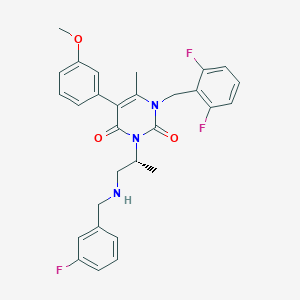 CHEMBL111933 CHEMBL111933 | C29H28F3N3O3 | 523.556 | 7 / 1 | 4.7 | No |
| 28499 | 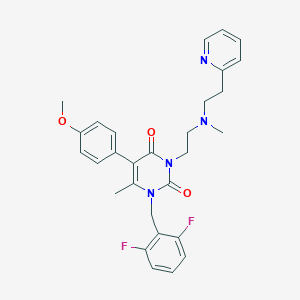 CHEMBL23900 CHEMBL23900 | C29H30F2N4O3 | 520.581 | 7 / 0 | 4.0 | No |
| 29936 | 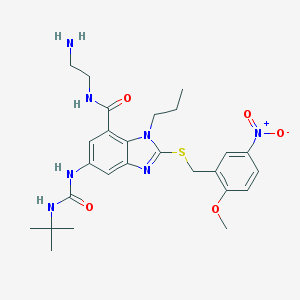 CHEMBL197317 CHEMBL197317 | C26H35N7O5S | 557.67 | 8 / 4 | 2.7 | No |
| 31012 | 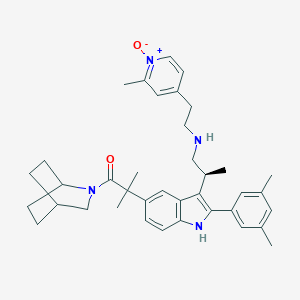 CHEMBL324654 CHEMBL324654 | C38H48N4O2 | 592.828 | 3 / 2 | 6.5 | No |
zhanglab![]() zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218