

You can:
| Name | Somatostatin receptor type 3 |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | SSTR3 |
| Synonym | SSR-28 SS3R SS3-R SS-3-R SRIF1C [ Show all ] |
| Disease | N/A |
| Length | 418 |
| Amino acid sequence | MDMLHPSSVSTTSEPENASSAWPPDATLGNVSAGPSPAGLAVSGVLIPLVYLVVCVVGLLGNSLVIYVVLRHTASPSVTNVYILNLALADELFMLGLPFLAAQNALSYWPFGSLMCRLVMAVDGINQFTSIFCLTVMSVDRYLAVVHPTRSARWRTAPVARTVSAAVWVASAVVVLPVVVFSGVPRGMSTCHMQWPEPAAAWRAGFIIYTAALGFFGPLLVICLCYLLIVVKVRSAGRRVWAPSCQRRRRSERRVTRMVVAVVALFVLCWMPFYVLNIVNVVCPLPEEPAFFGLYFLVVALPYANSCANPILYGFLSYRFKQGFRRVLLRPSRRVRSQEPTVGPPEKTEEEDEEEEDGEESREGGKGKEMNGRVSQITQPGTSGQERPPSRVASKEQQLLPQEASTGEKSSTMRISYL |
| UniProt | P32745 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | P32745 |
| 3D structure model | This predicted structure model is from GPCR-EXP P32745. |
| BioLiP | N/A |
| Therapeutic Target Database | N/A |
| ChEMBL | CHEMBL2028 |
| IUPHAR | 357 |
| DrugBank | BE0003529 |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 639 | 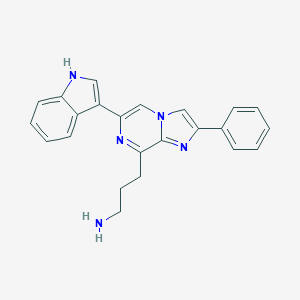 CHEMBL2112935 CHEMBL2112935 | C23H21N5 | 367.456 | 3 / 2 | 3.9 | Yes |
| 663 | 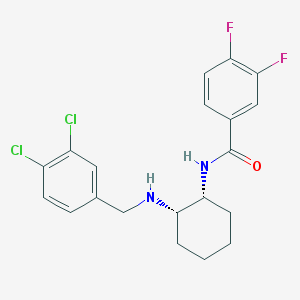 CHEMBL3287627 CHEMBL3287627 | C20H20Cl2F2N2O | 413.29 | 4 / 2 | 5.1 | No |
| 805 | 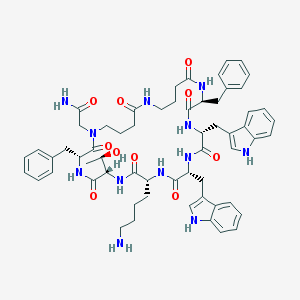 CHEMBL406000 CHEMBL406000 | C60H74N12O10 | 1123.33 | 11 / 12 | 3.1 | No |
| 1288 | 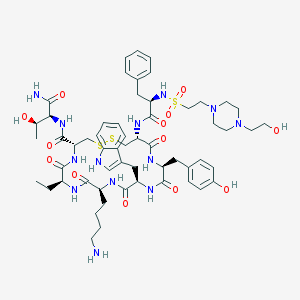 BIM 23197 BIM 23197 | C57H81N13O13S3 | 1252.53 | 19 / 14 | 0.0 | No |
| 3834 | 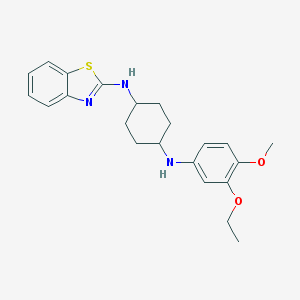 CHEMBL1079007 CHEMBL1079007 | C22H27N3O2S | 397.537 | 6 / 2 | 6.0 | No |
| 463391 | 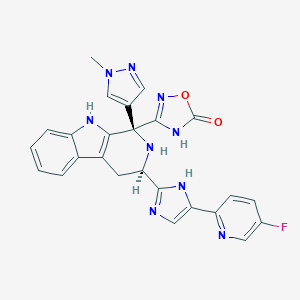 CHEMBL3582322 CHEMBL3582322 | C25H20FN9O2 | 497.494 | 8 / 4 | 1.5 | Yes |
| 4489 | 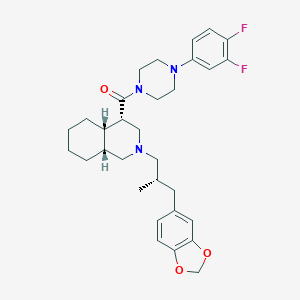 CHEMBL1076717 CHEMBL1076717 | C31H39F2N3O3 | 539.668 | 7 / 0 | 6.1 | No |
| 4490 | 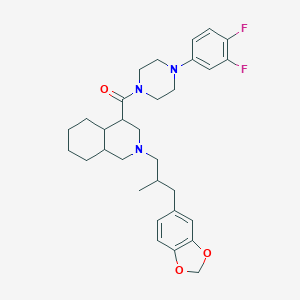 CHEMBL1081442 CHEMBL1081442 | C31H39F2N3O3 | 539.668 | 7 / 0 | 6.1 | No |
| 4678 | 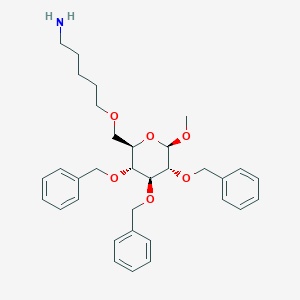 CHEMBL29102 CHEMBL29102 | C33H43NO6 | 549.708 | 7 / 1 | 4.2 | No |
| 6138 | 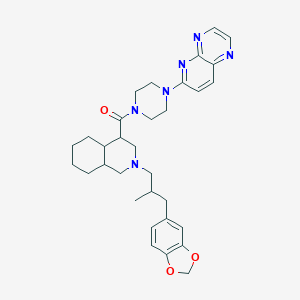 CHEMBL1076715 CHEMBL1076715 | C32H40N6O3 | 556.711 | 8 / 0 | 4.8 | No |
| 441929 | 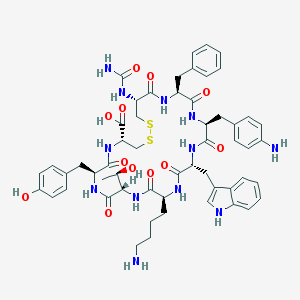 CHEMBL3350903 CHEMBL3350903 | C55H68N12O12S2 | 1153.34 | 16 / 15 | -0.2 | No |
| 441935 | 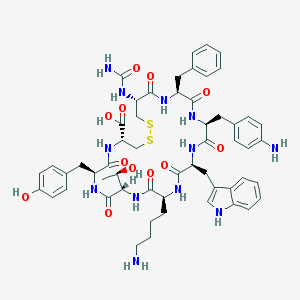 CHEMBL3350904 CHEMBL3350904 | C55H68N12O12S2 | 1153.34 | 16 / 15 | -0.2 | No |
| 6422 | 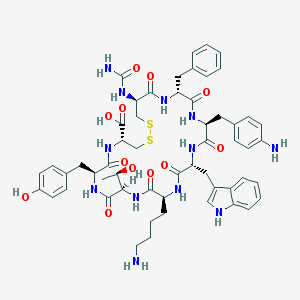 CHEMBL412183 CHEMBL412183 | C55H68N12O12S2 | 1153.34 | 16 / 15 | -0.2 | No |
| 6428 | 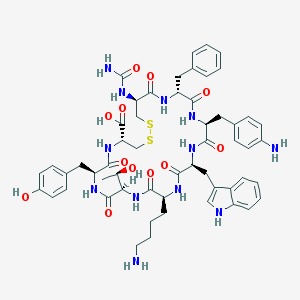 CHEMBL438546 CHEMBL438546 | C55H68N12O12S2 | 1153.34 | 16 / 15 | -0.2 | No |
| 7571 | 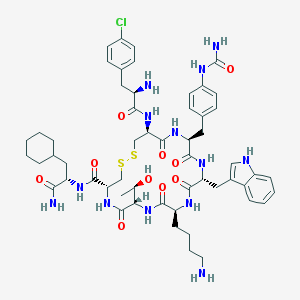 CHEMBL451932 CHEMBL451932 | C55H74ClN13O10S2 | 1176.85 | 14 / 14 | 3.1 | No |
| 8682 | 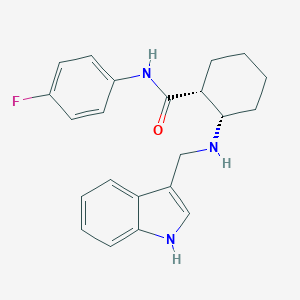 CHEMBL3287649 CHEMBL3287649 | C22H24FN3O | 365.452 | 3 / 3 | 4.4 | Yes |
| 8683 | 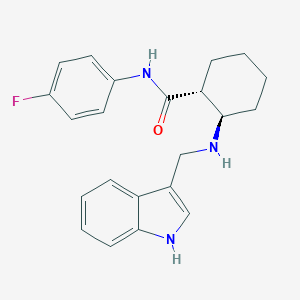 CHEMBL3287650 CHEMBL3287650 | C22H24FN3O | 365.452 | 3 / 3 | 4.4 | Yes |
| 9216 | 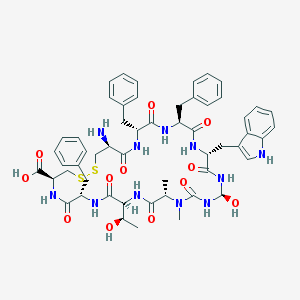 CHEMBL1161333 CHEMBL1161333 | C54H65N11O12S2 | 1124.3 | 15 / 13 | 0.3 | No |
| 9219 | 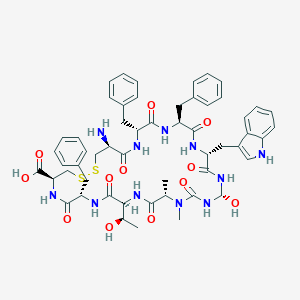 CHEMBL2093027 CHEMBL2093027 | C54H65N11O12S2 | 1124.3 | 15 / 13 | 0.3 | No |
| 9458 | 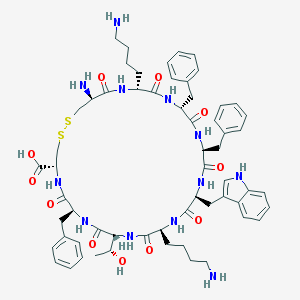 CHEMBL216992 CHEMBL216992 | C60H78N12O11S2 | 1207.48 | 16 / 14 | 0.6 | No |
| 10477 | 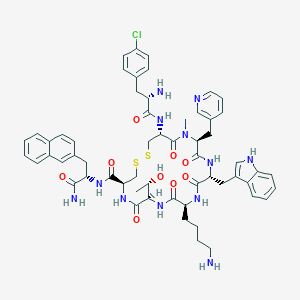 CHEMBL2369756 CHEMBL2369756 | C58H69ClN12O9S2 | 1177.83 | 14 / 11 | 3.8 | No |
| 10485 | 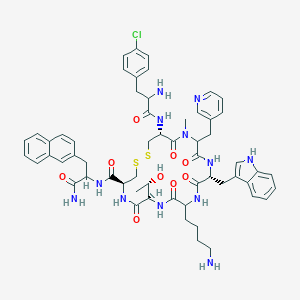 CHEMBL1794041 CHEMBL1794041 | C58H69ClN12O9S2 | 1177.83 | 14 / 11 | 3.8 | No |
| 10537 | 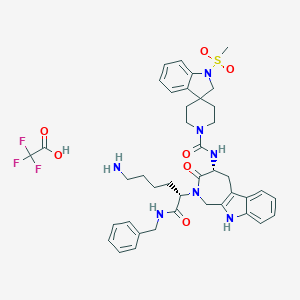 CHEMBL503379 CHEMBL503379 | C41H48F3N7O7S | 839.932 | 12 / 5 | N/A | No |
| 10928 | 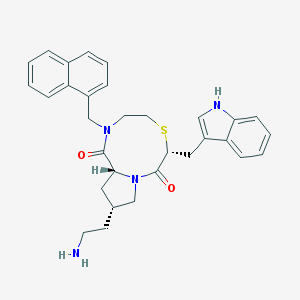 CHEMBL92545 CHEMBL92545 | C31H34N4O2S | 526.699 | 4 / 2 | 4.4 | No |
| 10934 | 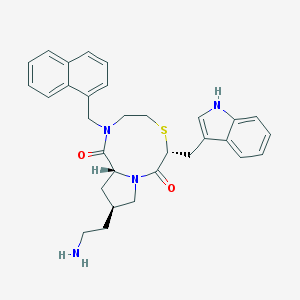 CHEMBL93618 CHEMBL93618 | C31H34N4O2S | 526.699 | 4 / 2 | 4.4 | No |
| 11589 | 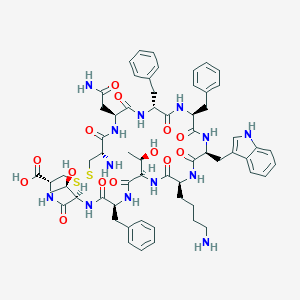 CHEMBL410047 CHEMBL410047 | C62H79N13O14S2 | 1294.51 | 18 / 16 | -1.0 | No |
| 11900 | 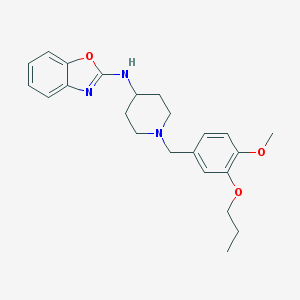 CHEMBL391951 CHEMBL391951 | C23H29N3O3 | 395.503 | 6 / 1 | 5.0 | Yes |
| 12004 | 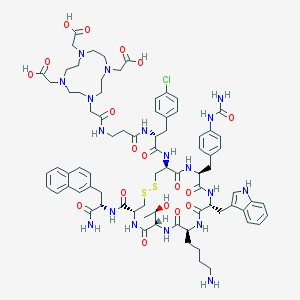 CHEMBL502077 CHEMBL502077 | C78H101ClN18O18S2 | 1678.34 | 25 / 18 | -5.4 | No |
| 12323 | 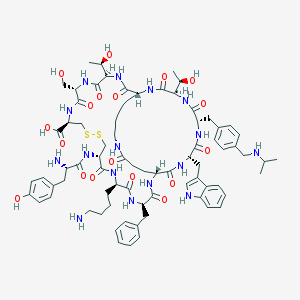 CHEMBL265839 CHEMBL265839 | C76H104N16O18S2 | 1593.88 | 23 / 21 | -0.9 | No |
| 13078 | 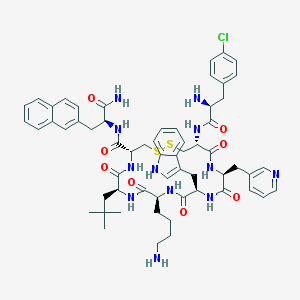 CHEMBL429166 CHEMBL429166 | C60H73ClN12O8S2 | 1189.89 | 13 / 11 | 5.4 | No |
| 13366 | 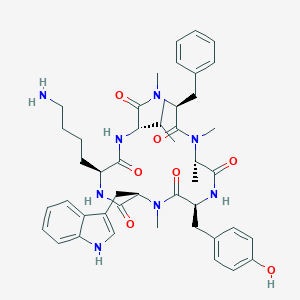 CHEMBL2011461 CHEMBL2011461 | C46H60N8O7 | 837.035 | 8 / 6 | 4.3 | No |
| 13544 | 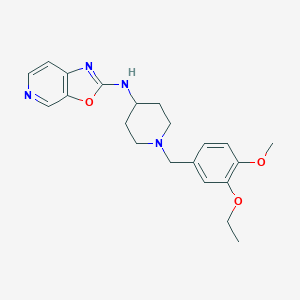 CHEMBL1077722 CHEMBL1077722 | C21H26N4O3 | 382.464 | 7 / 1 | 3.4 | Yes |
| 14430 | 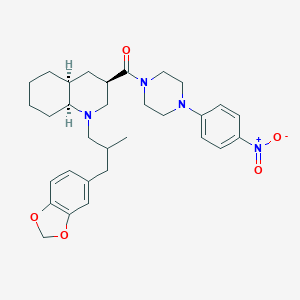 CHEMBL1076652 CHEMBL1076652 | C31H40N4O5 | 548.684 | 7 / 0 | 5.6 | No |
| 14433 | 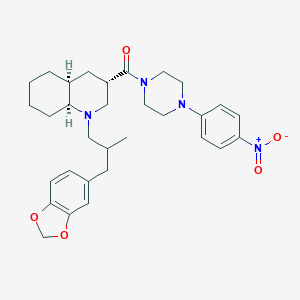 CHEMBL1076653 CHEMBL1076653 | C31H40N4O5 | 548.684 | 7 / 0 | 5.6 | No |
| 14435 | 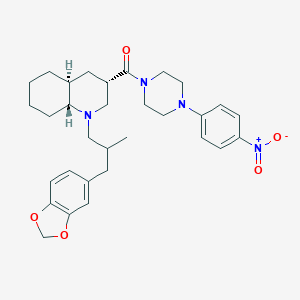 CHEMBL1076654 CHEMBL1076654 | C31H40N4O5 | 548.684 | 7 / 0 | 5.6 | No |
| 464679 | 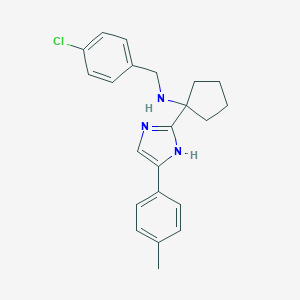 CHEMBL3605803 CHEMBL3605803 | C22H24ClN3 | 365.905 | 2 / 2 | 4.7 | Yes |
| 442228 | 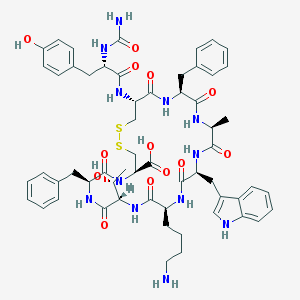 CHEMBL3350882 CHEMBL3350882 | C58H72N12O13S2 | 1209.41 | 16 / 15 | 0.2 | No |
| 14943 | 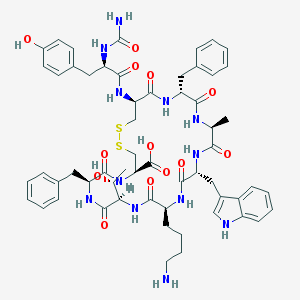 CHEMBL263292 CHEMBL263292 | C58H72N12O13S2 | 1209.41 | 16 / 15 | 0.2 | No |
| 14962 | 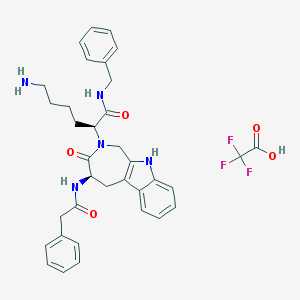 CHEMBL504838 CHEMBL504838 | C35H38F3N5O5 | 665.714 | 9 / 5 | N/A | No |
| 14964 | 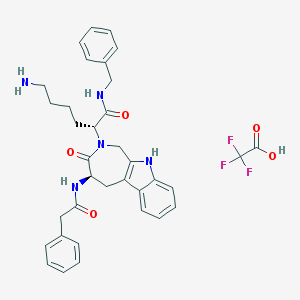 CHEMBL462020 CHEMBL462020 | C35H38F3N5O5 | 665.714 | 9 / 5 | N/A | No |
| 15213 | 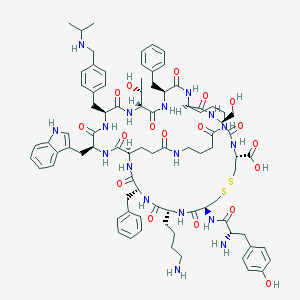 CHEMBL429164 CHEMBL429164 | C84H111N17O18S2 | 1711.03 | 23 / 21 | 0.0 | No |
| 15958 | 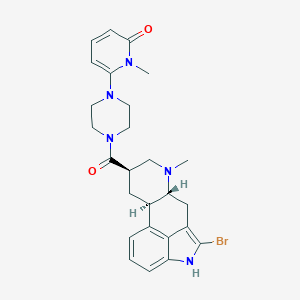 CHEMBL254500 CHEMBL254500 | C26H30BrN5O2 | 524.463 | 4 / 1 | 3.1 | No |
| 15961 | 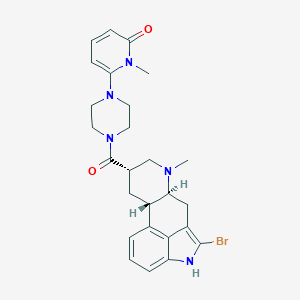 CHEMBL252231 CHEMBL252231 | C26H30BrN5O2 | 524.463 | 4 / 1 | 3.1 | No |
| 16297 | 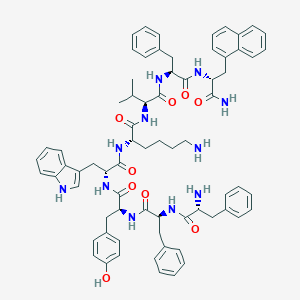 BDBM84629 BDBM84629 | C71H81N11O9 | 1232.5 | 11 / 12 | 8.1 | No |
| 17116 | 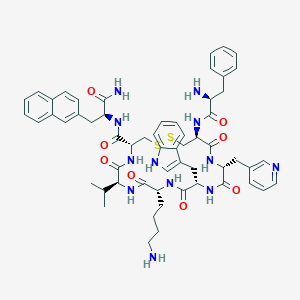 CHEMBL406260 CHEMBL406260 | C58H70N12O8S2 | 1127.39 | 13 / 11 | 4.0 | No |
| 442336 | 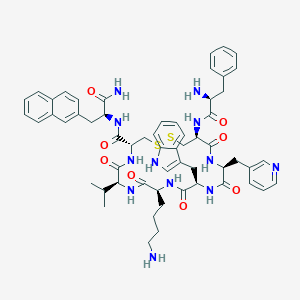 CHEMBL3349676 CHEMBL3349676 | C58H70N12O8S2 | 1127.39 | 13 / 11 | 4.0 | No |
| 17234 | 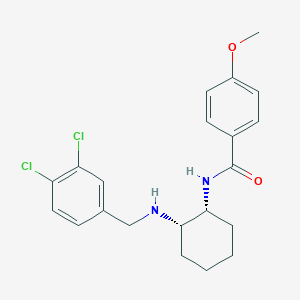 CHEMBL3287626 CHEMBL3287626 | C21H24Cl2N2O2 | 407.335 | 3 / 2 | 4.8 | Yes |
| 17586 | 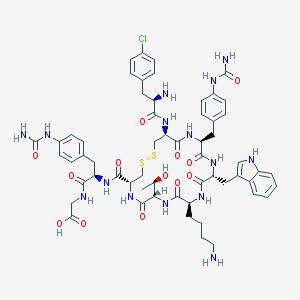 CHEMBL447177 CHEMBL447177 | C58H72ClN15O13S2 | 1286.88 | 17 / 17 | -1.7 | No |
| 18096 | 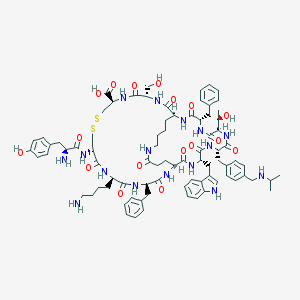 CHEMBL413888 CHEMBL413888 | C81H106N16O17S2 | 1639.95 | 22 / 20 | 0.7 | No |
| 442384 | 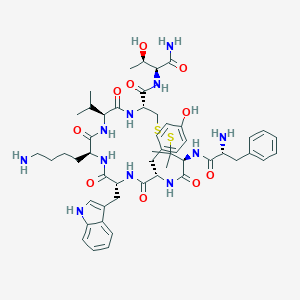 CHEMBL3349610 CHEMBL3349610 | C52H71N11O10S2 | 1074.33 | 14 / 13 | 1.9 | No |
| 18546 | 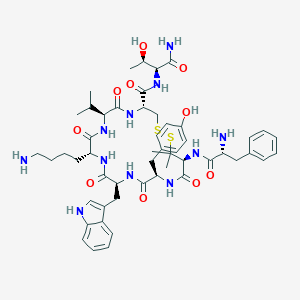 CHEMBL2370299 CHEMBL2370299 | C52H71N11O10S2 | 1074.33 | 14 / 13 | 1.9 | No |
| 18955 | 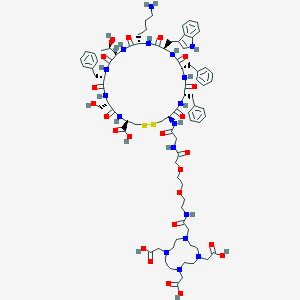 CHEMBL3122128 CHEMBL3122128 | C81H111N17O23S2 | 1754.99 | 30 / 19 | -9.1 | No |
| 19916 |  CHEMBL1223226 CHEMBL1223226 | C51H68N10O10 | 981.165 | 12 / 13 | 1.7 | No |
| 20633 |  CHEMBL414544 CHEMBL414544 | C57H69N9O11S2 | 1120.35 | 15 / 12 | 2.2 | No |
| 20642 |  CHEMBL2371100 CHEMBL2371100 | C57H69N9O11S2 | 1120.35 | 15 / 12 | 2.2 | No |
| 20644 |  CHEMBL2371108 CHEMBL2371108 | C57H69N9O11S2 | 1120.35 | 15 / 12 | 2.2 | No |
| 442483 | 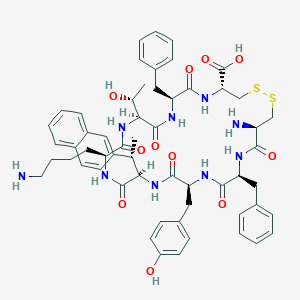 CHEMBL3350357 CHEMBL3350357 | C57H69N9O11S2 | 1120.35 | 15 / 12 | 2.2 | No |
| 20651 | 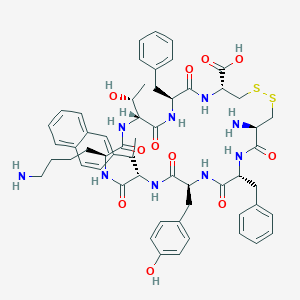 CHEMBL414272 CHEMBL414272 | C57H69N9O11S2 | 1120.35 | 15 / 12 | 2.2 | No |
| 20654 | 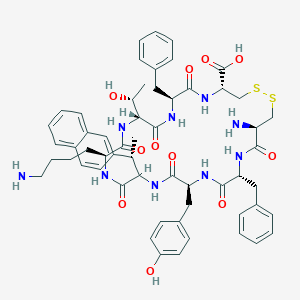 CHEMBL428281 CHEMBL428281 | C57H69N9O11S2 | 1120.35 | 15 / 12 | 2.2 | No |
| 20658 | 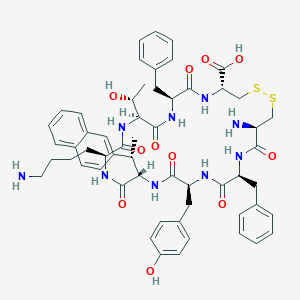 CHEMBL2111200 CHEMBL2111200 | C57H69N9O11S2 | 1120.35 | 15 / 12 | 2.2 | No |
| 20666 | 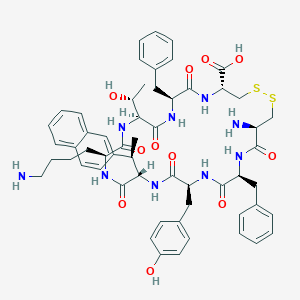 CHEMBL2111257 CHEMBL2111257 | C57H69N9O11S2 | 1120.35 | 15 / 12 | 2.2 | No |
| 20671 | 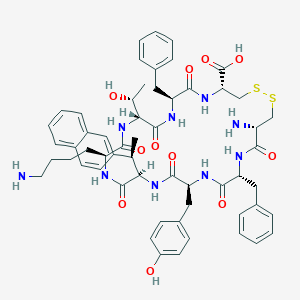 CHEMBL264046 CHEMBL264046 | C57H69N9O11S2 | 1120.35 | 15 / 12 | 2.2 | No |
| 20675 | 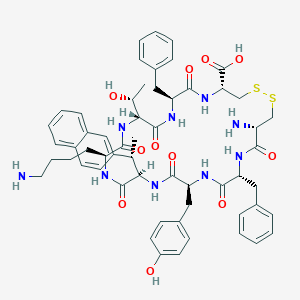 CHEMBL415582 CHEMBL415582 | C57H69N9O11S2 | 1120.35 | 15 / 12 | 2.2 | No |
| 20984 | 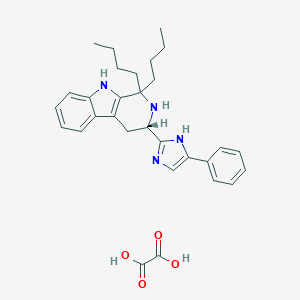 CHEMBL1788167 CHEMBL1788167 | C30H36N4O4 | 516.642 | 6 / 5 | N/A | No |
| 522076 | 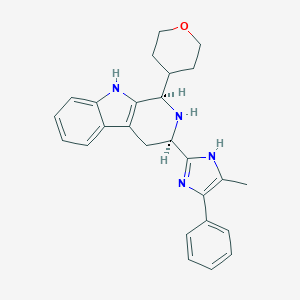 CHEMBL3775846 CHEMBL3775846 | C26H28N4O | 412.537 | 3 / 3 | 3.9 | Yes |
| 21950 | 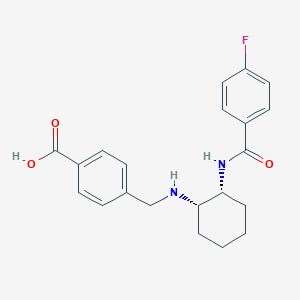 CHEMBL3287620 CHEMBL3287620 | C21H23FN2O3 | 370.424 | 5 / 3 | 0.9 | Yes |
| 465722 | 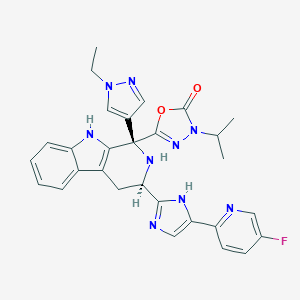 CHEMBL3582340 CHEMBL3582340 | C29H28FN9O2 | 553.602 | 8 / 3 | 2.8 | No |
| 522303 | 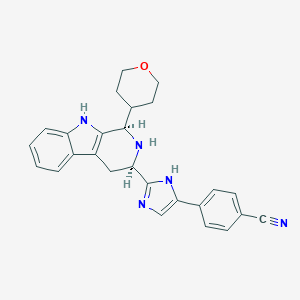 CHEMBL3774861 CHEMBL3774861 | C26H25N5O | 423.52 | 4 / 3 | 3.2 | Yes |
| 24682 | 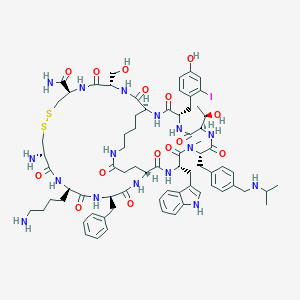 CHEMBL387005 CHEMBL387005 | C73H99IN16O15S2 | 1631.72 | 20 / 18 | 1.9 | No |
| 25080 | 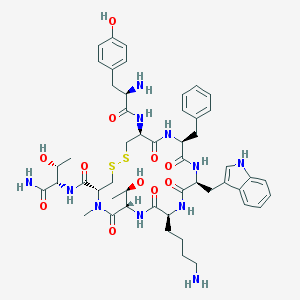 CHEMBL384836 CHEMBL384836 | C50H67N11O11S2 | 1062.27 | 15 / 13 | 0.4 | No |
| 25591 | 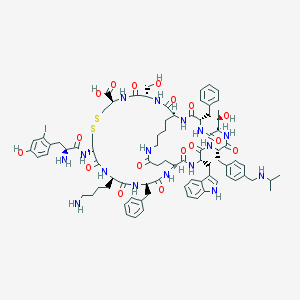 CHEMBL440074 CHEMBL440074 | C81H105IN16O17S2 | 1765.85 | 22 / 20 | 1.4 | No |
| 26163 | 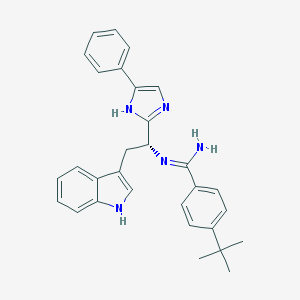 CHEMBL176730 CHEMBL176730 | C30H31N5 | 461.613 | 2 / 3 | 6.2 | No |
| 26265 | 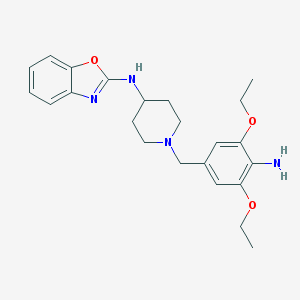 CHEMBL1078449 CHEMBL1078449 | C23H30N4O3 | 410.518 | 7 / 2 | 4.2 | Yes |
| 26750 | 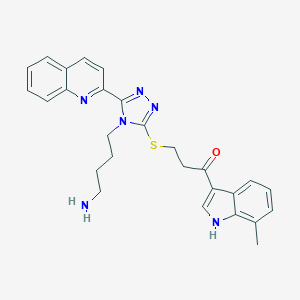 CHEMBL434159 CHEMBL434159 | C27H28N6OS | 484.622 | 6 / 2 | 3.7 | Yes |
| 442730 | 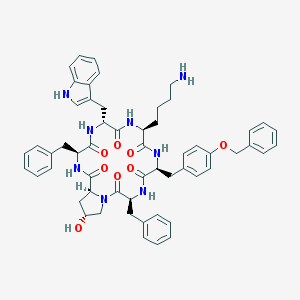 CHEMBL3349599 CHEMBL3349599 | C56H62N8O8 | 975.16 | 9 / 8 | 5.5 | No |
| 466266 | 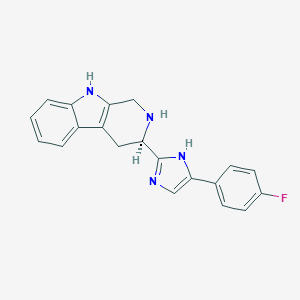 CHEMBL3582307 CHEMBL3582307 | C20H17FN4 | 332.382 | 3 / 3 | 3.0 | Yes |
| 27598 | 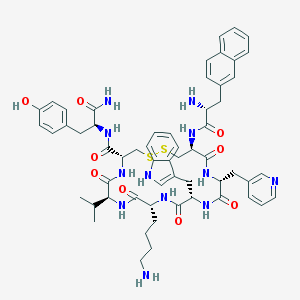 CHEMBL409754 CHEMBL409754 | C58H70N12O9S2 | 1143.39 | 14 / 12 | 3.6 | No |
| 442742 | 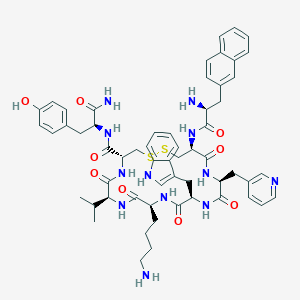 CHEMBL3349669 CHEMBL3349669 | C58H70N12O9S2 | 1143.39 | 14 / 12 | 3.6 | No |
| 28079 | 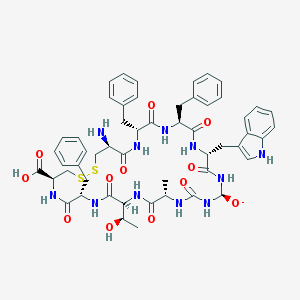 sst4-selective analogue 17 sst4-selective analogue 17 | C53H62N11O12S2- | 1109.26 | 15 / 13 | 1.2 | No |
| 28629 | 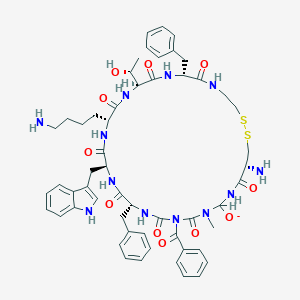 BDBM50136762 BDBM50136762 | C55H67N12O11S2- | 1136.33 | 15 / 11 | 3.7 | No |
| 28805 | 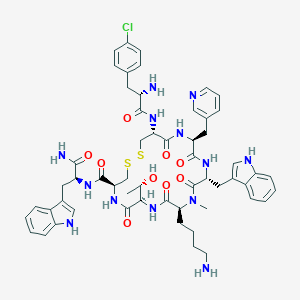 CHEMBL2369735 CHEMBL2369735 | C56H68ClN13O9S2 | 1166.81 | 14 / 12 | 2.6 | No |
| 28807 | 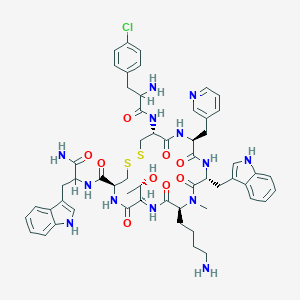 CHEMBL1793861 CHEMBL1793861 | C56H68ClN13O9S2 | 1166.81 | 14 / 12 | 2.6 | No |
| 466466 | 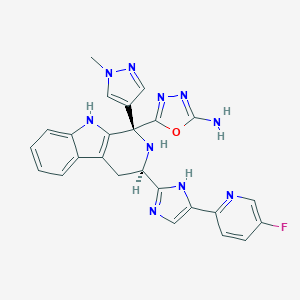 CHEMBL3582330 CHEMBL3582330 | C25H21FN10O | 496.51 | 9 / 4 | 0.9 | Yes |
| 29011 | 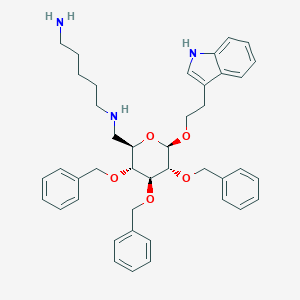 CHEMBL282129 CHEMBL282129 | C42H51N3O5 | 677.886 | 7 / 3 | 6.0 | No |
| 522450 | 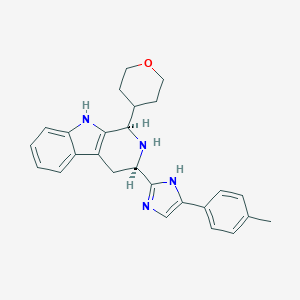 CHEMBL3774791 CHEMBL3774791 | C26H28N4O | 412.537 | 3 / 3 | 3.8 | Yes |
| 442852 | 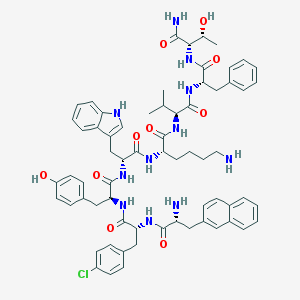 CHEMBL3349508 CHEMBL3349508 | C66H78ClN11O10 | 1220.87 | 12 / 13 | 6.6 | No |
| 30544 | 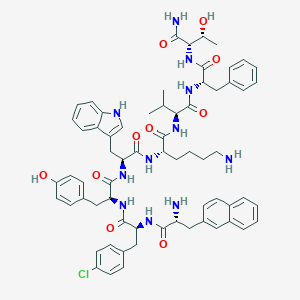 CHEMBL1793962 CHEMBL1793962 | C66H78ClN11O10 | 1220.87 | 12 / 13 | 6.6 | No |
| 442858 | 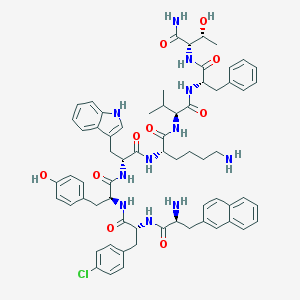 CHEMBL3349506 CHEMBL3349506 | C66H78ClN11O10 | 1220.87 | 12 / 13 | 6.6 | No |
| 30546 | 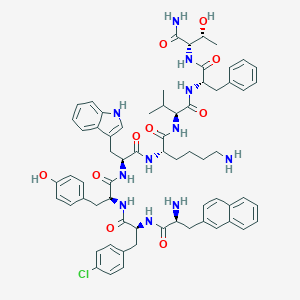 CHEMBL1793960 CHEMBL1793960 | C66H78ClN11O10 | 1220.87 | 12 / 13 | 6.6 | No |
| 30917 | 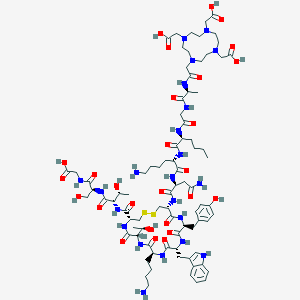 CHEMBL3122125 CHEMBL3122125 | C82H126N22O27S2 | 1916.16 | 35 / 26 | -14.6 | No |
| 33864 | 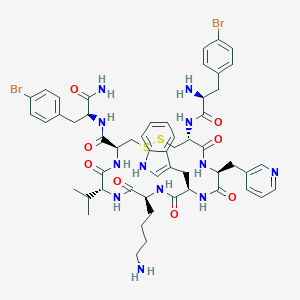 CHEMBL437448 CHEMBL437448 | C54H66Br2N12O8S2 | 1235.13 | 13 / 11 | 4.1 | No |
| 33973 | 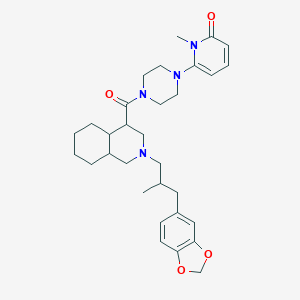 CHEMBL1079799 CHEMBL1079799 | C31H42N4O4 | 534.701 | 6 / 0 | 4.6 | No |
| 467227 | 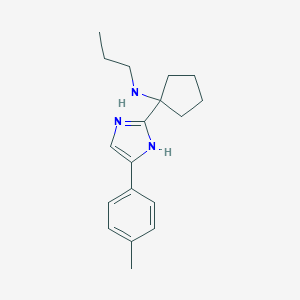 CHEMBL3605799 CHEMBL3605799 | C18H25N3 | 283.419 | 2 / 2 | 3.4 | Yes |
| 36269 | 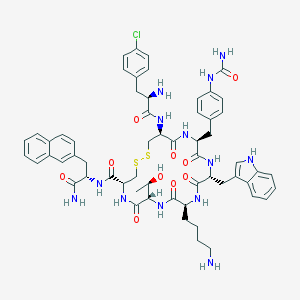 CHEMBL502219 CHEMBL502219 | C59H70ClN13O10S2 | 1220.86 | 14 / 14 | 3.2 | No |
| 36290 | 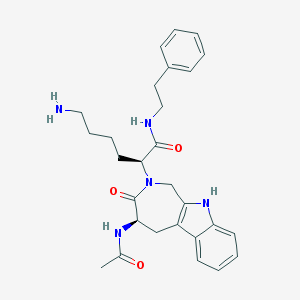 CHEMBL442605 CHEMBL442605 | C28H35N5O3 | 489.62 | 4 / 4 | 2.3 | Yes |
| 37387 | 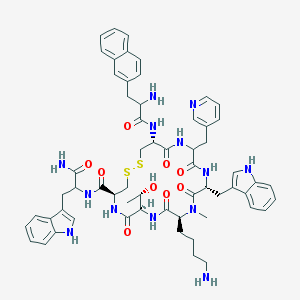 CHEMBL1794043 CHEMBL1794043 | C60H71N13O9S2 | 1182.43 | 14 / 12 | 3.3 | No |
| 37394 | 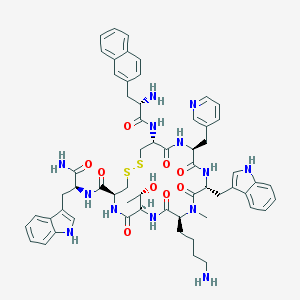 CHEMBL2369758 CHEMBL2369758 | C60H71N13O9S2 | 1182.43 | 14 / 12 | 3.3 | No |
| 37571 | 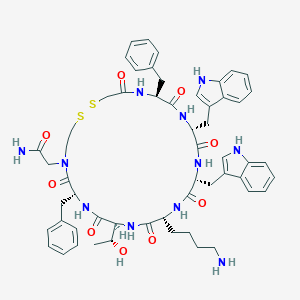 CHEMBL408471 CHEMBL408471 | C56H67N11O9S2 | 1102.34 | 12 / 11 | 3.9 | No |
| 37655 | 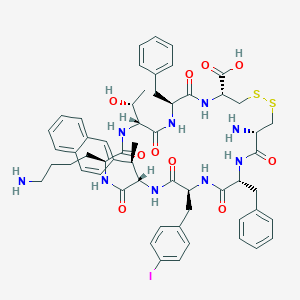 CHEMBL414446 CHEMBL414446 | C57H68IN9O10S2 | 1230.25 | 14 / 11 | 3.2 | No |
| 37658 | 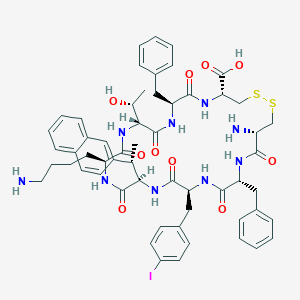 CHEMBL407649 CHEMBL407649 | C57H68IN9O10S2 | 1230.25 | 14 / 11 | 3.2 | No |
zhanglab![]() zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218