

You can:
| Name | D(3) dopamine receptor |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | DRD3 |
| Synonym | dopaminergic receptor D3 D3R D3 receptor dopamine D3 receptor |
| Disease | Unspecified Emesis; Gastric motility disorder Female sexual dysfunction Male sexual disorders Psychotic disorders [ Show all ] |
| Length | 400 |
| Amino acid sequence | MASLSQLSSHLNYTCGAENSTGASQARPHAYYALSYCALILAIVFGNGLVCMAVLKERALQTTTNYLVVSLAVADLLVATLVMPWVVYLEVTGGVWNFSRICCDVFVTLDVMMCTASILNLCAISIDRYTAVVMPVHYQHGTGQSSCRRVALMITAVWVLAFAVSCPLLFGFNTTGDPTVCSISNPDFVIYSSVVSFYLPFGVTVLVYARIYVVLKQRRRKRILTRQNSQCNSVRPGFPQQTLSPDPAHLELKRYYSICQDTALGGPGFQERGGELKREEKTRNSLSPTIAPKLSLEVRKLSNGRLSTSLKLGPLQPRGVPLREKKATQMVAIVLGAFIVCWLPFFLTHVLNTHCQTCHVSPELYSATTWLGYVNSALNPVIYTTFNIEFRKAFLKILSC |
| UniProt | P35462 |
| Protein Data Bank | 3pbl |
| GPCR-HGmod model | P35462 |
| 3D structure model | This structure is from PDB ID 3pbl. |
| BioLiP | BL0191566, BL0191567 |
| Therapeutic Target Database | T02551 |
| ChEMBL | CHEMBL234 |
| IUPHAR | 216 |
| DrugBank | BE0000581 |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 83 | 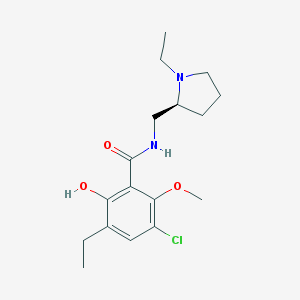 Eticlopride Eticlopride | C17H25ClN2O3 | 340.848 | 4 / 2 | 3.1 | Yes |
| 95 | 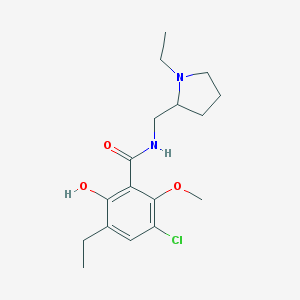 AC1O8PYY AC1O8PYY | C17H25ClN2O3 | 340.848 | 4 / 2 | 3.1 | Yes |
| 124 | 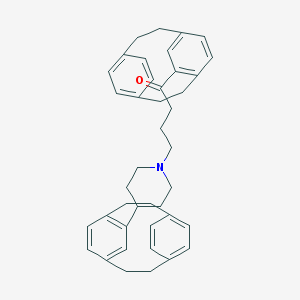 CHEMBL1256170 CHEMBL1256170 | C41H45NO | 567.817 | 2 / 0 | 9.2 | No |
| 223 | 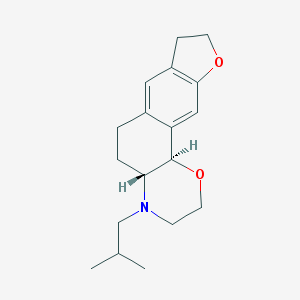 CHEMBL169322 CHEMBL169322 | C18H25NO2 | 287.403 | 3 / 0 | 3.2 | Yes |
| 392 | 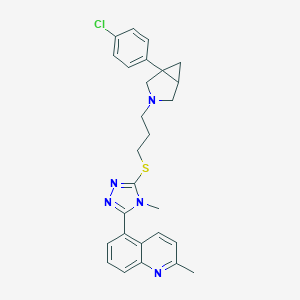 CHEMBL1080117 CHEMBL1080117 | C27H28ClN5S | 490.066 | 5 / 0 | 5.5 | No |
| 535913 | 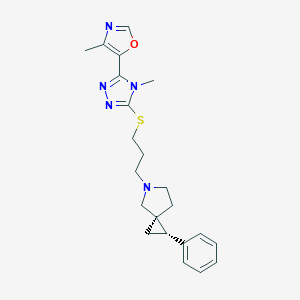 CHEMBL3958494 CHEMBL3958494 | C22H27N5OS | 409.552 | 6 / 0 | 3.3 | Yes |
| 535914 | 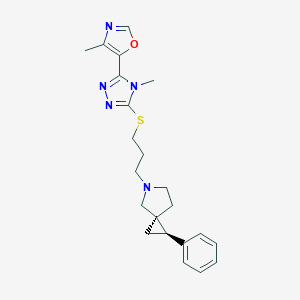 CHEMBL3913062 CHEMBL3913062 | C22H27N5OS | 409.552 | 6 / 0 | 3.3 | Yes |
| 535916 | 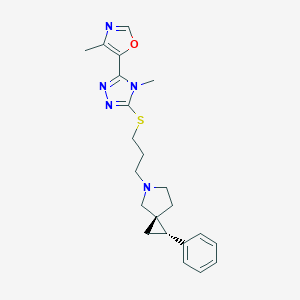 CHEMBL3983937 CHEMBL3983937 | C22H27N5OS | 409.552 | 6 / 0 | 3.3 | Yes |
| 535923 | 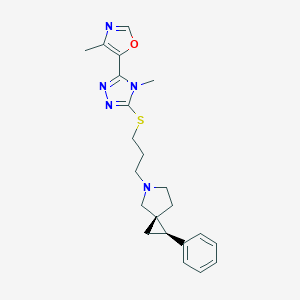 CHEMBL3901211 CHEMBL3901211 | C22H27N5OS | 409.552 | 6 / 0 | 3.3 | Yes |
| 473 | 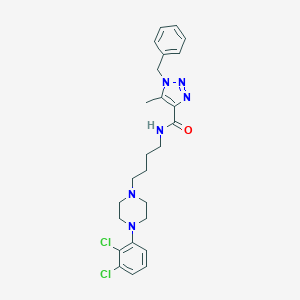 CHEMBL373873 CHEMBL373873 | C25H30Cl2N6O | 501.456 | 5 / 1 | 4.9 | No |
| 631 | 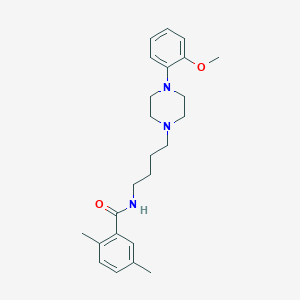 CHEMBL1258494 CHEMBL1258494 | C24H33N3O2 | 395.547 | 4 / 1 | 4.2 | Yes |
| 521454 | 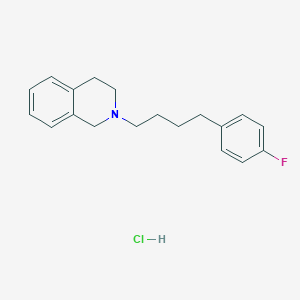 CHEMBL3818128 CHEMBL3818128 | C19H23ClFN | 319.848 | 2 / 1 | N/A | N/A |
| 984 |  CHEMBL221681 CHEMBL221681 | C24H28BrN3O3 | 486.41 | 5 / 1 | 4.9 | Yes |
| 1022 |  CHEMBL2205816 CHEMBL2205816 | C18H20N2O | 280.371 | 3 / 1 | 1.8 | Yes |
| 1046 |  CHEMBL270853 CHEMBL270853 | C30H34N6OS | 526.703 | 6 / 0 | 4.4 | No |
| 1061 |  CHEMBL2420770 CHEMBL2420770 | C15H17FN2S | 276.373 | 4 / 0 | 3.2 | Yes |
| 1095 | 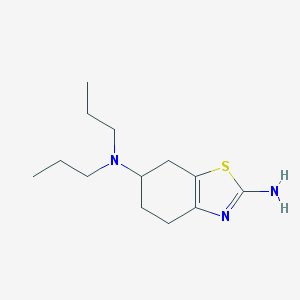 CHEMBL105512 CHEMBL105512 | C13H23N3S | 253.408 | 4 / 1 | 3.2 | Yes |
| 1258 | 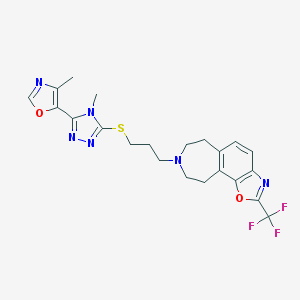 CHEMBL256659 CHEMBL256659 | C22H23F3N6O2S | 492.521 | 11 / 0 | 3.9 | No |
| 1281 | 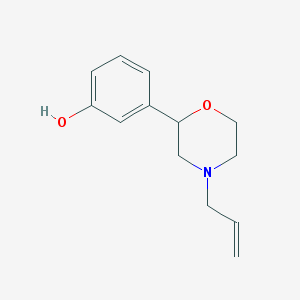 CHEMBL398480 CHEMBL398480 | C13H17NO2 | 219.284 | 3 / 1 | 2.1 | Yes |
| 1452 | 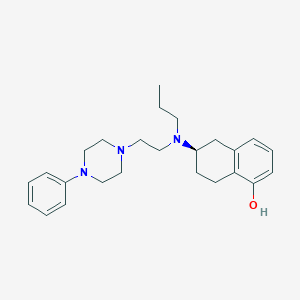 CHEMBL457025 CHEMBL457025 | C25H35N3O | 393.575 | 4 / 1 | 4.8 | Yes |
| 1457 |  CHEMBL457024 CHEMBL457024 | C25H35N3O | 393.575 | 4 / 1 | 4.8 | Yes |
| 1459 |  CHEMBL250552 CHEMBL250552 | C25H35N3O | 393.575 | 4 / 1 | 4.8 | Yes |
| 1503 |  CHEMBL1257447 CHEMBL1257447 | C22H27N5OS | 409.552 | 6 / 0 | 3.5 | Yes |
| 1582 |  CHEMBL207543 CHEMBL207543 | C22H26Cl2N6 | 445.392 | 5 / 0 | 4.3 | Yes |
| 521488 |  CHEMBL3781035 CHEMBL3781035 | C25H33NO4 | 411.542 | 5 / 1 | 5.2 | No |
| 1657 |  CHEMBL210461 CHEMBL210461 | C31H35Cl2N3O | 536.541 | 3 / 1 | 7.3 | No |
| 535956 |  CHEMBL3940897 CHEMBL3940897 | C23H25F3N6S | 474.55 | 9 / 0 | 3.3 | Yes |
| 535959 |  CHEMBL3939830 CHEMBL3939830 | C23H25F3N6S | 474.55 | 9 / 0 | 3.3 | Yes |
| 1739 | 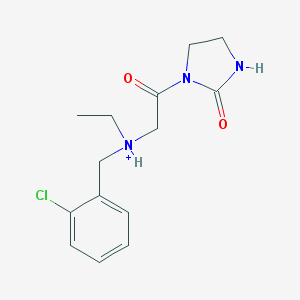 (2-Chlorophenyl)methyl-ethyl-[2-oxo-2-(2-oxoimidazolidin-1-yl)ethyl]azanium (2-Chlorophenyl)methyl-ethyl-[2-oxo-2-(2-oxoimidazolidin-1-yl)ethyl]azanium | C14H19ClN3O2+ | 296.775 | 2 / 2 | 1.7 | Yes |
| 1811 | 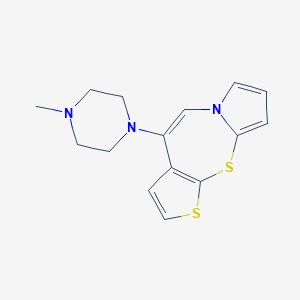 CHEMBL180319 CHEMBL180319 | C15H17N3S2 | 303.442 | 4 / 0 | 3.0 | Yes |
| 1839 | 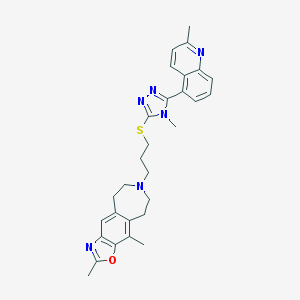 CHEMBL272694 CHEMBL272694 | C29H32N6OS | 512.676 | 7 / 0 | 5.6 | No |
| 441744 | 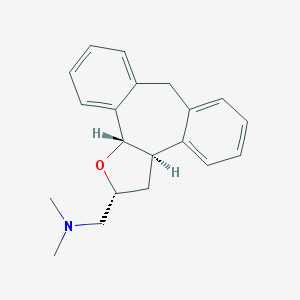 CHEMBL329566 CHEMBL329566 | C20H23NO | 293.41 | 2 / 0 | 3.6 | Yes |
| 1927 |  CHEMBL371352 CHEMBL371352 | C20H23NO | 293.41 | 2 / 0 | 3.6 | Yes |
| 2099 |  CHEMBL558838 CHEMBL558838 | C21H28N6O | 380.496 | 6 / 1 | 2.9 | Yes |
| 2191 |  CHEMBL458002 CHEMBL458002 | C29H28ClN5O2 | 514.026 | 5 / 2 | 4.3 | No |
| 2278 |  1-(cyclohexylmethyl)pyrrolidine 1-(cyclohexylmethyl)pyrrolidine | C11H21N | 167.296 | 1 / 0 | 3.1 | Yes |
| 441835 | 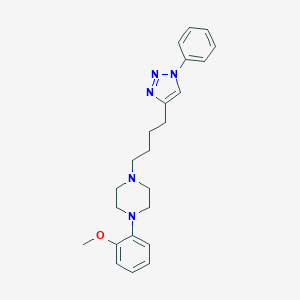 CHEMBL3353895 CHEMBL3353895 | C23H29N5O | 391.519 | 5 / 0 | 4.0 | Yes |
| 521537 | 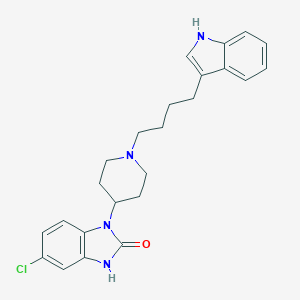 CHEMBL3818805 CHEMBL3818805 | C24H27ClN4O | 422.957 | 2 / 2 | 4.8 | Yes |
| 3146 | 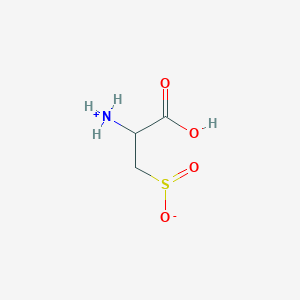 AC1N0YGA AC1N0YGA | C3H7NO4S | 153.152 | 5 / 2 | -4.7 | Yes |
| 3284 | 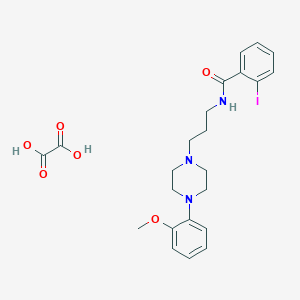 CHEMBL263058 CHEMBL263058 | C23H28IN3O6 | 569.396 | 8 / 3 | N/A | No |
| 463297 |  MLS001217465 MLS001217465 | C23H24N2O7 | 440.452 | 8 / 2 | N/A | N/A |
| 521542 |  CHEMBL3742207 CHEMBL3742207 | C22H26ClN5O | 411.934 | 6 / 1 | 5.1 | No |
| 3467 |  CHEMBL80711 CHEMBL80711 | C26H32N4O6 | 496.564 | 9 / 3 | N/A | N/A |
| 3574 |  CHEMBL238915 CHEMBL238915 | C21H24ClN3O3S | 433.951 | 5 / 0 | 3.6 | Yes |
| 521544 | 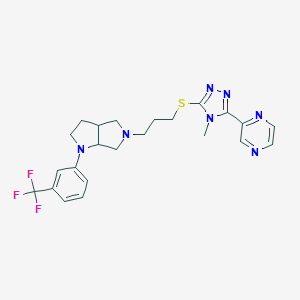 CHEMBL3774425 CHEMBL3774425 | C23H26F3N7S | 489.565 | 10 / 0 | 3.2 | Yes |
| 463330 | 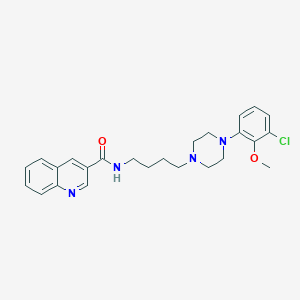 CHEMBL3597637 CHEMBL3597637 | C25H29ClN4O2 | 452.983 | 5 / 1 | 4.4 | Yes |
| 463340 | 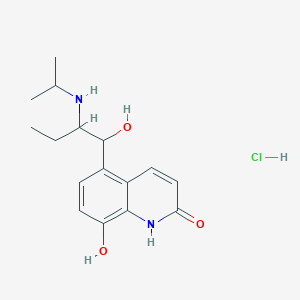 procaterol hydrochloride procaterol hydrochloride | C16H23ClN2O3 | 326.821 | 4 / 5 | N/A | N/A |
| 3796 | 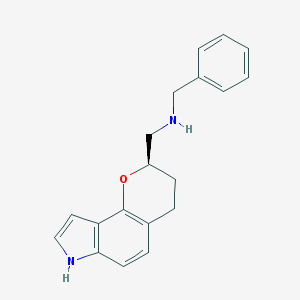 CHEMBL60185 CHEMBL60185 | C19H20N2O | 292.382 | 2 / 2 | 3.5 | Yes |
| 3839 |  CHEMBL1257214 CHEMBL1257214 | C23H28FN5S2 | 457.63 | 7 / 0 | 4.6 | Yes |
| 3894 |  CHEMBL239917 CHEMBL239917 | C22H26ClN3O | 383.92 | 3 / 0 | 3.9 | Yes |
| 3928 |  CHEMBL1079480 CHEMBL1079480 | C23H25F3N6S | 474.55 | 9 / 0 | 3.4 | Yes |
| 4243 |  CHEMBL301475 CHEMBL301475 | C25H26F3N3O4S | 521.555 | 8 / 2 | 4.8 | No |
| 4270 | 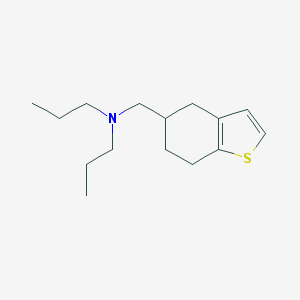 CHEMBL104135 CHEMBL104135 | C15H25NS | 251.432 | 2 / 0 | 4.4 | Yes |
| 4323 | 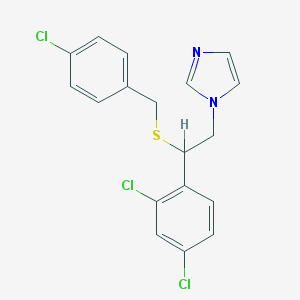 sulconazole sulconazole | C18H15Cl3N2S | 397.742 | 2 / 0 | 6.1 | No |
| 463416 | 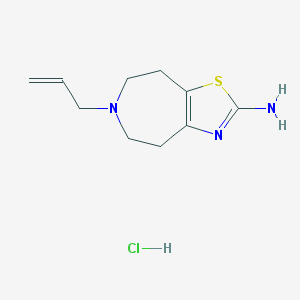 talipexole hydrochloride talipexole hydrochloride | C10H16ClN3S | 245.769 | 4 / 2 | N/A | N/A |
| 4477 | 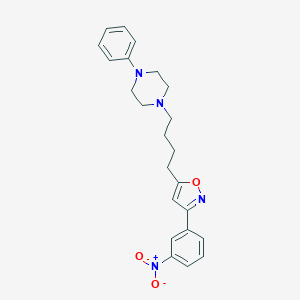 CHEMBL538977 CHEMBL538977 | C23H26N4O3 | 406.486 | 6 / 0 | 4.5 | Yes |
| 4508 |  CHEMBL497604 CHEMBL497604 | C24H26N4O2 | 402.498 | 5 / 1 | 4.0 | Yes |
| 521582 |  CHEMBL3797876 CHEMBL3797876 | C17H21NO5 | 319.357 | 6 / 4 | 1.6 | Yes |
| 521584 |  CHEMBL3799593 CHEMBL3799593 | C17H21NO5 | 319.357 | 6 / 4 | 1.6 | Yes |
| 4608 |  CHEMBL1258493 CHEMBL1258493 | C24H31N5O2S | 453.605 | 7 / 0 | 4.0 | Yes |
| 4662 | 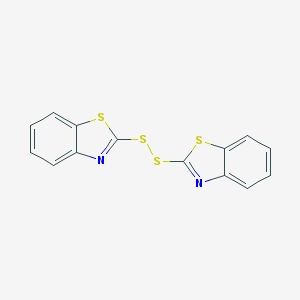 120-78-5 120-78-5 | C14H8N2S4 | 332.472 | 6 / 0 | 5.6 | No |
| 4688 | 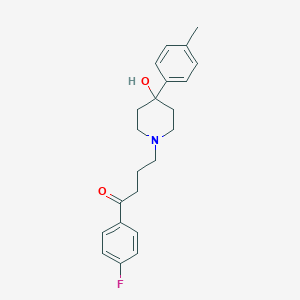 Moperone Moperone | C22H26FNO2 | 355.453 | 4 / 1 | 3.0 | Yes |
| 4827 | 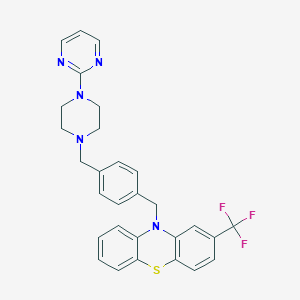 CHEMBL232206 CHEMBL232206 | C29H26F3N5S | 533.617 | 9 / 0 | 6.2 | No |
| 4892 | 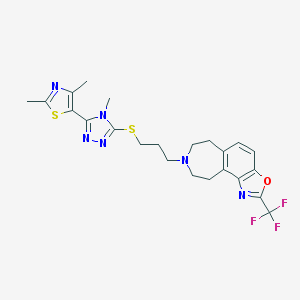 CHEMBL270376 CHEMBL270376 | C23H25F3N6OS2 | 522.609 | 11 / 0 | 4.9 | No |
| 4956 |  5HT6-ligand-1 5HT6-ligand-1 | C20H22BrN3O2S | 448.379 | 4 / 0 | 3.6 | Yes |
| 5034 |  CHEMBL211135 CHEMBL211135 | C17H17Cl2N5 | 362.258 | 4 / 0 | 3.4 | Yes |
| 521608 |  CHEMBL3764191 CHEMBL3764191 | C21H23F3N4OS2 | 468.557 | 9 / 0 | 4.0 | Yes |
| 5112 |  CHEMBL338938 CHEMBL338938 | C25H35N3O2 | 409.574 | 3 / 1 | 4.7 | Yes |
| 5235 |  CHEMBL196744 CHEMBL196744 | C21H24Cl2FN3O | 424.341 | 4 / 1 | 4.9 | Yes |
| 5256 |  SB-258719 SB-258719 | C18H30N2O2S | 338.51 | 4 / 0 | 3.5 | Yes |
| 5367 |  CHEMBL396560 CHEMBL396560 | C21H24N2O | 320.436 | 2 / 1 | 4.3 | Yes |
| 5847 |  CHEMBL93594 CHEMBL93594 | C15H23NO3S | 297.413 | 4 / 0 | 2.9 | Yes |
| 5883 |  CHEMBL402422 CHEMBL402422 | C22H26N6O2S | 438.55 | 8 / 0 | 3.4 | Yes |
| 5923 |  CHEMBL24809 CHEMBL24809 | C22H25N3 | 331.463 | 2 / 0 | 4.3 | Yes |
| 5994 |  CHEMBL129931 CHEMBL129931 | C18H23NO | 269.388 | 2 / 1 | 4.1 | Yes |
| 6086 |  CHEMBL551964 CHEMBL551964 | C27H25N5O2 | 451.53 | 7 / 0 | 4.3 | Yes |
| 6090 |  CID 10072510 CID 10072510 | C27H25N5O2 | 451.53 | 7 / 0 | 4.3 | Yes |
| 6125 |  CHEMBL1689001 CHEMBL1689001 | C25H31BrFN3O4 | 536.442 | 7 / 1 | 4.1 | No |
| 6263 |  CHEMBL1173436 CHEMBL1173436 | C22H24N4O2 | 376.46 | 3 / 2 | 2.9 | Yes |
| 6272 |  CHEMBL76237 CHEMBL76237 | C19H22N2O3S | 358.456 | 4 / 0 | 3.5 | Yes |
| 6301 | 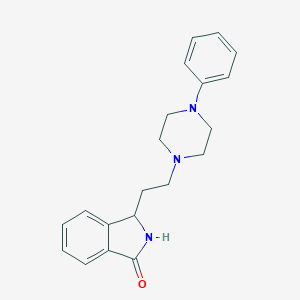 CHEMBL34239 CHEMBL34239 | C20H23N3O | 321.424 | 3 / 1 | 2.8 | Yes |
| 6637 | 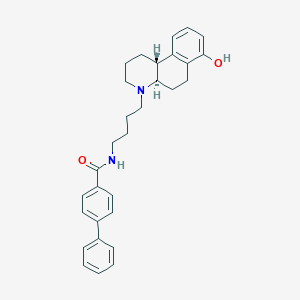 CHEMBL419706 CHEMBL419706 | C30H34N2O2 | 454.614 | 3 / 2 | 5.9 | No |
| 6639 | 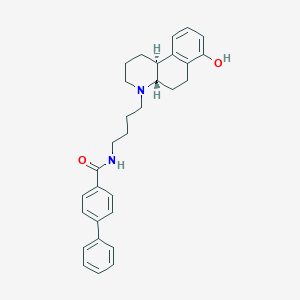 CHEMBL99198 CHEMBL99198 | C30H34N2O2 | 454.614 | 3 / 2 | 5.9 | No |
| 6740 | 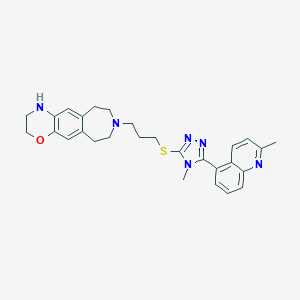 CHEMBL408169 CHEMBL408169 | C28H32N6OS | 500.665 | 7 / 1 | 4.7 | No |
| 521647 |  CHEMBL3775737 CHEMBL3775737 | C23H32FN5OS | 445.601 | 7 / 0 | 3.2 | Yes |
| 6931 |  CHEMBL424458 CHEMBL424458 | C26H34F3NO | 433.559 | 5 / 0 | 7.5 | No |
| 441972 |  CHEMBL3358498 CHEMBL3358498 | C24H27FN4O | 406.505 | 5 / 1 | 4.2 | Yes |
| 463731 |  CHEMBL3633757 CHEMBL3633757 | C26H24BrNO4 | 494.385 | 5 / 0 | 5.4 | No |
| 441981 | 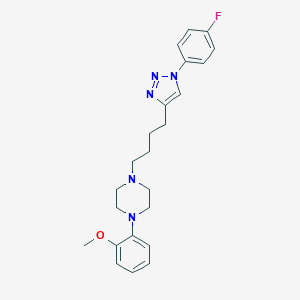 CHEMBL3353904 CHEMBL3353904 | C23H28FN5O | 409.509 | 6 / 0 | 4.1 | Yes |
| 7073 | 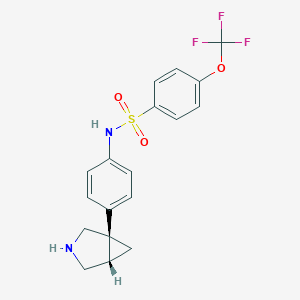 CHEMBL1224701 CHEMBL1224701 | C18H17F3N2O3S | 398.4 | 8 / 2 | 3.4 | Yes |
| 7129 | 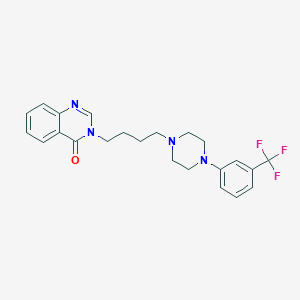 CHEMBL398813 CHEMBL398813 | C23H25F3N4O | 430.475 | 7 / 0 | 4.1 | Yes |
| 463788 | 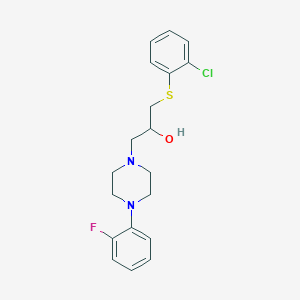 1-[(2-chlorophenyl)sulfanyl]-3-[4-(2-fluorophenyl)piperazino]-2-propanol 1-[(2-chlorophenyl)sulfanyl]-3-[4-(2-fluorophenyl)piperazino]-2-propanol | C19H22ClFN2OS | 380.906 | 5 / 1 | 4.2 | Yes |
| 7691 | 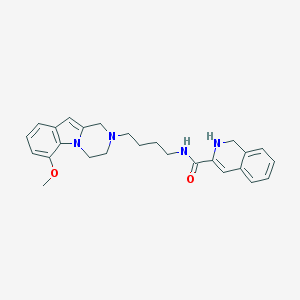 CID 44581146 CID 44581146 | C26H30N4O2 | 430.552 | 4 / 2 | 3.5 | Yes |
| 7891 | 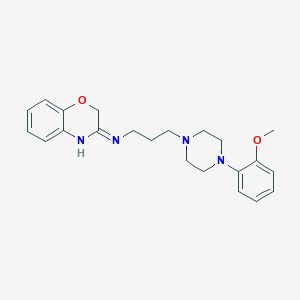 CHEMBL80711 CHEMBL80711 | C22H28N4O2 | 380.492 | 5 / 1 | 2.9 | Yes |
| 8110 | 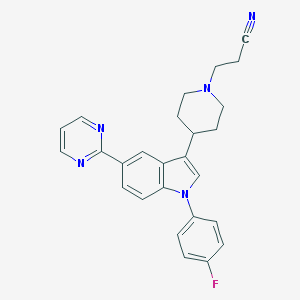 CHEMBL317333 CHEMBL317333 | C26H24FN5 | 425.511 | 5 / 0 | 4.0 | Yes |
| 8325 | 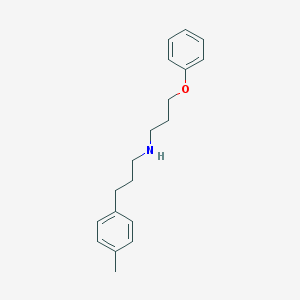 CHEMBL334989 CHEMBL334989 | C19H25NO | 283.415 | 2 / 1 | 4.5 | Yes |
| 8377 | 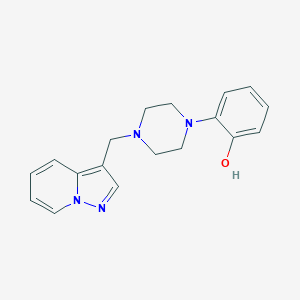 CHEMBL272602 CHEMBL272602 | C18H20N4O | 308.385 | 4 / 1 | 2.1 | Yes |
| 521685 | 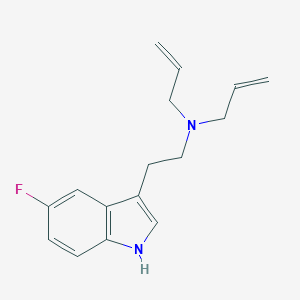 CHEMBL3754496 CHEMBL3754496 | C16H19FN2 | 258.34 | 2 / 1 | 2.9 | Yes |
| 8697 | 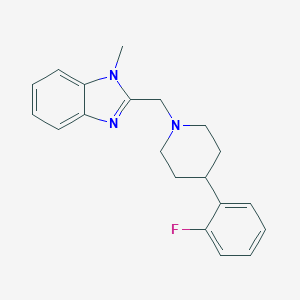 CHEMBL607689 CHEMBL607689 | C20H22FN3 | 323.415 | 3 / 0 | 3.6 | Yes |
| 8786 | 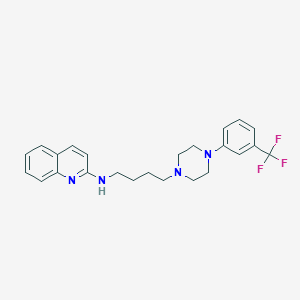 CHEMBL1223747 CHEMBL1223747 | C24H27F3N4 | 428.503 | 7 / 1 | 5.7 | No |
zhanglab![]() zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218