

You can:
| Name | Metabotropic glutamate receptor 5 |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | GRM5 |
| Synonym | mGlu5 receptor glutamate receptor GPRC1E mGluR5 |
| Disease | Central nervous system disease Chronic neuropathic pain Fragile X syndrome Major depressive disorder; GERD; Chronic neuropathic pain Autism [ Show all ] |
| Length | 1212 |
| Amino acid sequence | MVLLLILSVLLLKEDVRGSAQSSERRVVAHMPGDIIIGALFSVHHQPTVDKVHERKCGAVREQYGIQRVEAMLHTLERINSDPTLLPNITLGCEIRDSCWHSAVALEQSIEFIRDSLISSEEEEGLVRCVDGSSSSFRSKKPIVGVIGPGSSSVAIQVQNLLQLFNIPQIAYSATSMDLSDKTLFKYFMRVVPSDAQQARAMVDIVKRYNWTYVSAVHTEGNYGESGMEAFKDMSAKEGICIAHSYKIYSNAGEQSFDKLLKKLTSHLPKARVVACFCEGMTVRGLLMAMRRLGLAGEFLLLGSDGWADRYDVTDGYQREAVGGITIKLQSPDVKWFDDYYLKLRPETNHRNPWFQEFWQHRFQCRLEGFPQENSKYNKTCNSSLTLKTHHVQDSKMGFVINAIYSMAYGLHNMQMSLCPGYAGLCDAMKPIDGRKLLESLMKTNFTGVSGDTILFDENGDSPGRYEIMNFKEMGKDYFDYINVGSWDNGELKMDDDEVWSKKSNIIRSVCSEPCEKGQIKVIRKGEVSCCWTCTPCKENEYVFDEYTCKACQLGSWPTDDLTGCDLIPVQYLRWGDPEPIAAVVFACLGLLATLFVTVVFIIYRDTPVVKSSSRELCYIILAGICLGYLCTFCLIAKPKQIYCYLQRIGIGLSPAMSYSALVTKTNRIARILAGSKKKICTKKPRFMSACAQLVIAFILICIQLGIIVALFIMEPPDIMHDYPSIREVYLICNTTNLGVVTPLGYNGLLILSCTFYAFKTRNVPANFNEAKYIAFTMYTTCIIWLAFVPIYFGSNYKIITMCFSVSLSATVALGCMFVPKVYIILAKPERNVRSAFTTSTVVRMHVGDGKSSSAASRSSSLVNLWKRRGSSGETLRYKDRRLAQHKSEIECFTPKGSMGNGGRATMSSSNGKSVTWAQNEKSSRGQHLWQRLSIHINKKENPNQTAVIKPFPKSTESRGLGAGAGAGGSAGGVGATGGAGCAGAGPGGPESPDAGPKALYDVAEAEEHFPAPARPRSPSPISTLSHRAGSASRTDDDVPSLHSEPVARSSSSQGSLMEQISSVVTRFTANISELNSMMLSTAAPSPGVGAPLCSSYLIPKEIQLPTTMTTFAEIQPLPAIEVTGGAQPAAGAQAAGDAARESPAAGPEAAAAKPDLEELVALTPPSPFRDSVDSGSTTPNSPVSESALCIPSSPKYDTLIIRDYTQSSSSL |
| UniProt | P41594 |
| Protein Data Bank | 4oo9, 5cgc, 5cgd, 6ffh, 6ffi, 6n4x, 6n4y, 6n50, 6n51, 3lmk |
| GPCR-HGmod model | N/A |
| 3D structure model | This structure is from PDB ID 4oo9. |
| BioLiP | BL0176927,BL0176931, BL0176928,BL0176929,BL0176930, BL0281199, BL0322076, BL0322077, BL0407724, BL0438693, BL0438694, BL0438695,BL0438696,BL0438697, BL0407725, BL0438698,BL0438699 |
| Therapeutic Target Database | T99347 |
| ChEMBL | CHEMBL3227 |
| IUPHAR | 293 |
| DrugBank | BE0001192 |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 521449 | 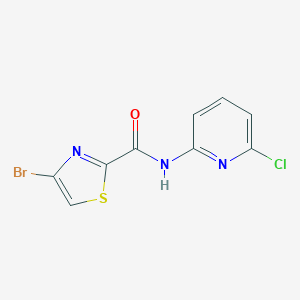 CHEMBL3739885 CHEMBL3739885 | C9H5BrClN3OS | 318.573 | 4 / 1 | 3.4 | Yes |
| 648 | 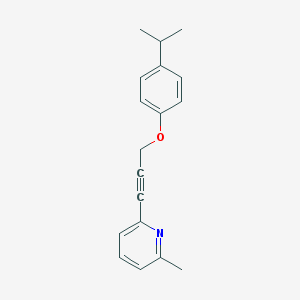 CHEMBL215957 CHEMBL215957 | C18H19NO | 265.356 | 2 / 0 | 4.3 | Yes |
| 459240 | 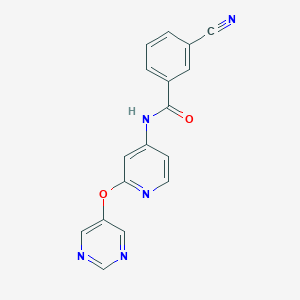 CHEMBL3655617 CHEMBL3655617 | C17H11N5O2 | 317.308 | 6 / 1 | 1.5 | Yes |
| 905 | 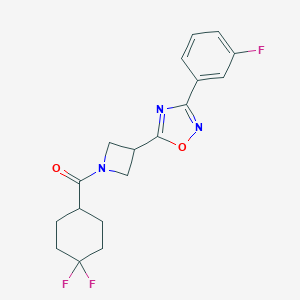 CHEMBL2151617 CHEMBL2151617 | C18H18F3N3O2 | 365.356 | 7 / 0 | 3.1 | Yes |
| 1086 | 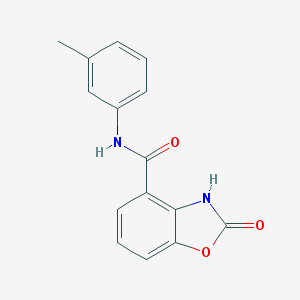 CHEMBL246041 CHEMBL246041 | C15H12N2O3 | 268.272 | 3 / 2 | 2.3 | Yes |
| 521473 | 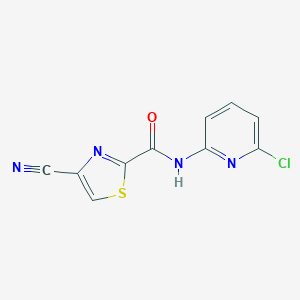 CHEMBL3742147 CHEMBL3742147 | C10H5ClN4OS | 264.687 | 5 / 1 | 2.4 | Yes |
| 535945 | 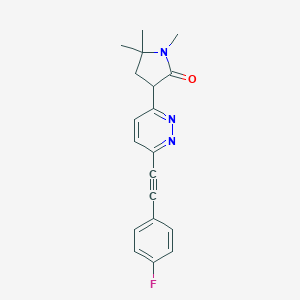 CHEMBL3982481 CHEMBL3982481 | C19H18FN3O | 323.371 | 4 / 0 | 2.2 | Yes |
| 1207 | 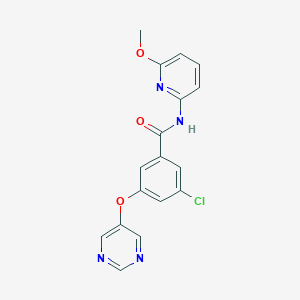 SCHEMBL6918188 SCHEMBL6918188 | C17H13ClN4O3 | 356.766 | 6 / 1 | 2.8 | Yes |
| 1219 | 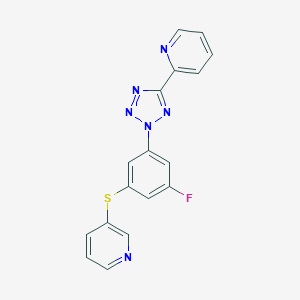 CHEMBL184047 CHEMBL184047 | C17H11FN6S | 350.375 | 7 / 0 | 3.5 | Yes |
| 1538 | 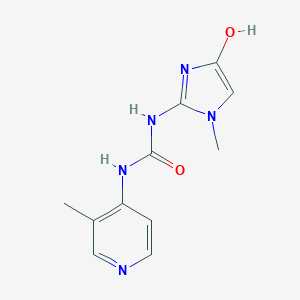 BDBM50201676 BDBM50201676 | C11H13N5O2 | 247.258 | 4 / 3 | 0.2 | Yes |
| 521501 | 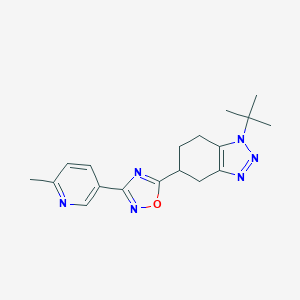 CHEMBL3736163 CHEMBL3736163 | C18H22N6O | 338.415 | 6 / 0 | 2.3 | Yes |
| 2101 | 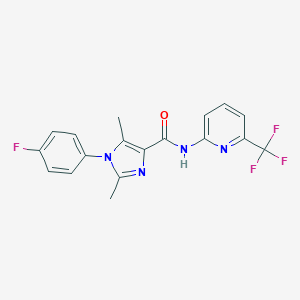 CHEMBL2346732 CHEMBL2346732 | C18H14F4N4O | 378.331 | 7 / 1 | 3.9 | Yes |
| 2195 | 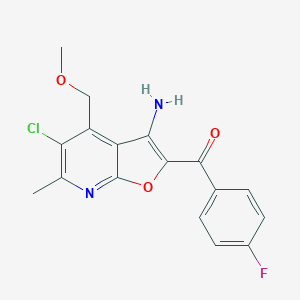 CHEMBL1784103 CHEMBL1784103 | C17H14ClFN2O3 | 348.758 | 6 / 1 | 3.9 | Yes |
| 2931 | 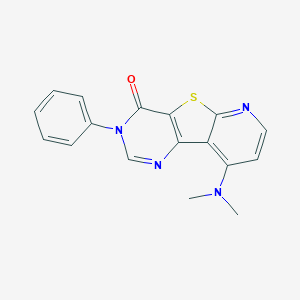 CHEMBL224615 CHEMBL224615 | C17H14N4OS | 322.386 | 5 / 0 | 3.2 | Yes |
| 441856 | 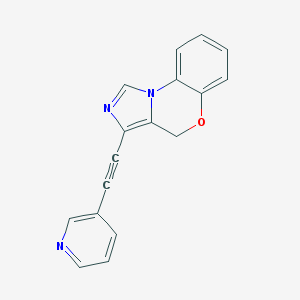 CHEMBL3410212 CHEMBL3410212 | C17H11N3O | 273.295 | 3 / 0 | 2.2 | Yes |
| 536022 | 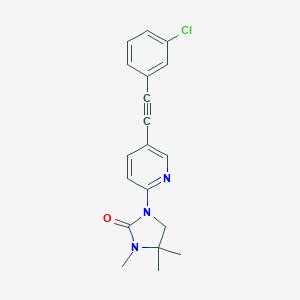 SCHEMBL2520315 SCHEMBL2520315 | C19H18ClN3O | 339.823 | 2 / 0 | 3.6 | Yes |
| 4033 | 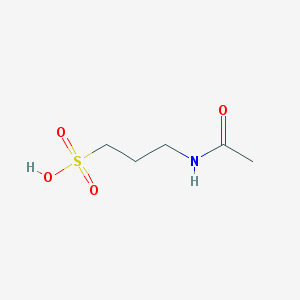 Acamprosate Acamprosate | C5H11NO4S | 181.206 | 4 / 2 | -1.2 | Yes |
| 4245 | 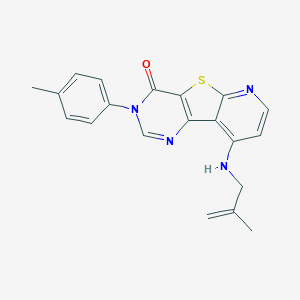 CHEMBL1783868 CHEMBL1783868 | C20H18N4OS | 362.451 | 5 / 1 | 4.7 | Yes |
| 4404 | 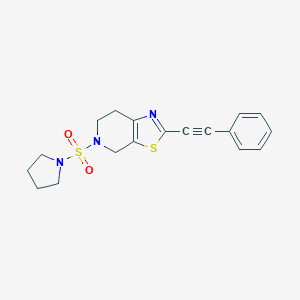 CHEMBL2403653 CHEMBL2403653 | C18H19N3O2S2 | 373.489 | 6 / 0 | 2.6 | Yes |
| 5150 | 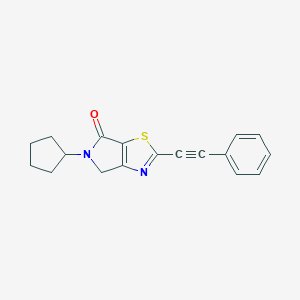 CHEMBL2403667 CHEMBL2403667 | C18H16N2OS | 308.399 | 3 / 0 | 3.7 | Yes |
| 5302 | 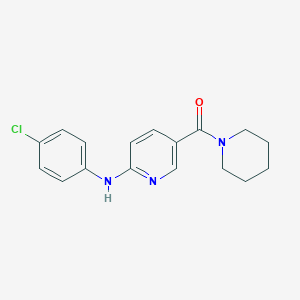 CHEMBL595943 CHEMBL595943 | C17H18ClN3O | 315.801 | 3 / 1 | 3.7 | Yes |
| 5810 | 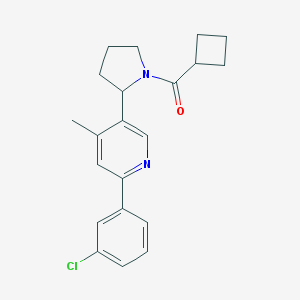 CHEMBL1808881 CHEMBL1808881 | C21H23ClN2O | 354.878 | 2 / 0 | 4.3 | Yes |
| 459277 | 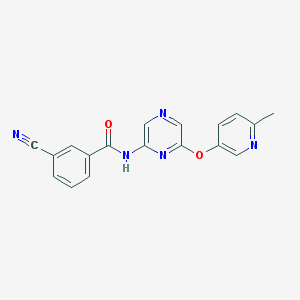 CHEMBL3655594 CHEMBL3655594 | C18H13N5O2 | 331.335 | 6 / 1 | 1.9 | Yes |
| 6345 | 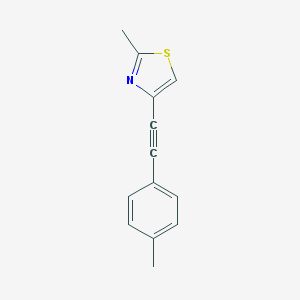 CHEMBL185813 CHEMBL185813 | C13H11NS | 213.298 | 2 / 0 | 3.8 | Yes |
| 441959 | 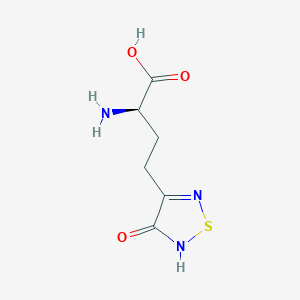 CHEMBL334842 CHEMBL334842 | C6H9N3O3S | 203.216 | 6 / 3 | -3.4 | Yes |
| 441961 | 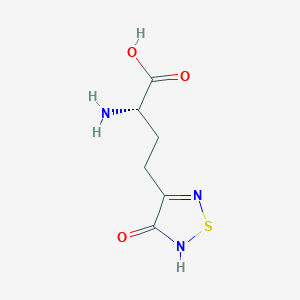 CHEMBL131922 CHEMBL131922 | C6H9N3O3S | 203.216 | 6 / 3 | -3.4 | Yes |
| 6751 | 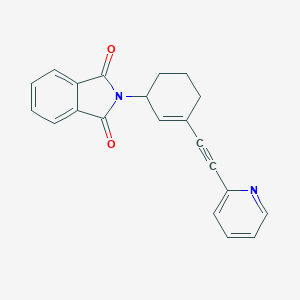 CHEMBL2113099 CHEMBL2113099 | C21H16N2O2 | 328.371 | 3 / 0 | 2.9 | Yes |
| 6815 | 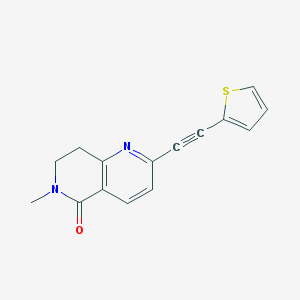 CHEMBL1779857 CHEMBL1779857 | C15H12N2OS | 268.334 | 3 / 0 | 2.2 | Yes |
| 536111 | 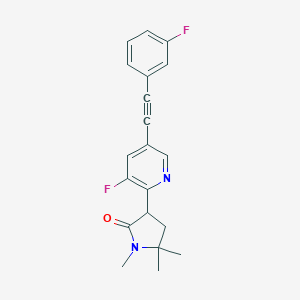 CHEMBL3939809 CHEMBL3939809 | C20H18F2N2O | 340.374 | 4 / 0 | 3.3 | Yes |
| 536112 | 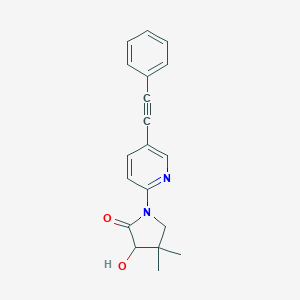 SCHEMBL2518446 SCHEMBL2518446 | C19H18N2O2 | 306.365 | 3 / 1 | 2.9 | Yes |
| 7026 | 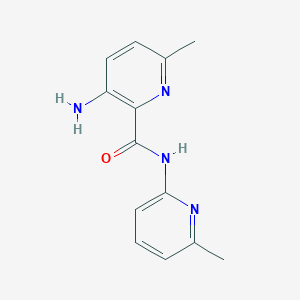 CHEMBL181424 CHEMBL181424 | C13H14N4O | 242.282 | 4 / 2 | 2.0 | Yes |
| 536141 | 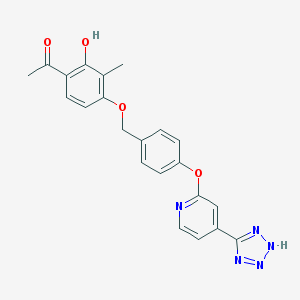 CHEMBL3899832 CHEMBL3899832 | C22H19N5O4 | 417.425 | 8 / 2 | 3.6 | Yes |
| 7203 | 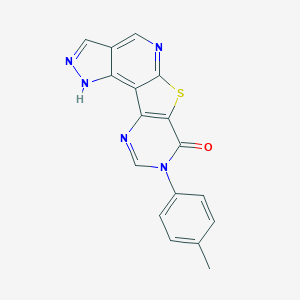 CHEMBL238262 CHEMBL238262 | C17H11N5OS | 333.369 | 5 / 1 | 3.1 | Yes |
| 536161 | 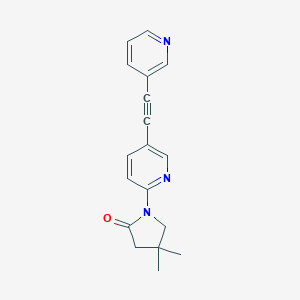 SCHEMBL2520891 SCHEMBL2520891 | C18H17N3O | 291.354 | 3 / 0 | 2.4 | Yes |
| 7429 | 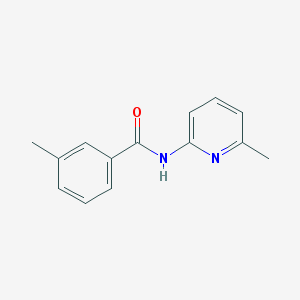 3-methyl-N-(6-methylpyridin-2-yl)benzamide 3-methyl-N-(6-methylpyridin-2-yl)benzamide | C14H14N2O | 226.279 | 2 / 1 | 2.8 | Yes |
| 7791 | 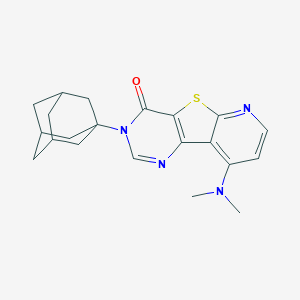 CHEMBL224322 CHEMBL224322 | C21H24N4OS | 380.51 | 5 / 0 | 4.3 | Yes |
| 7817 | 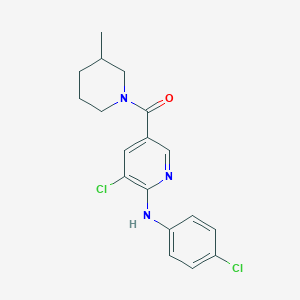 CHEMBL594296 CHEMBL594296 | C18H19Cl2N3O | 364.27 | 3 / 1 | 4.7 | Yes |
| 7836 | 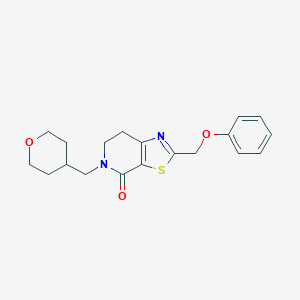 CHEMBL2426602 CHEMBL2426602 | C19H22N2O3S | 358.456 | 5 / 0 | 2.9 | Yes |
| 536173 | 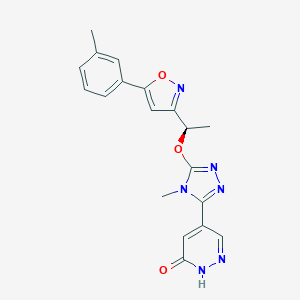 SCHEMBL2959176 SCHEMBL2959176 | C19H18N6O3 | 378.392 | 7 / 1 | 1.2 | Yes |
| 7880 | 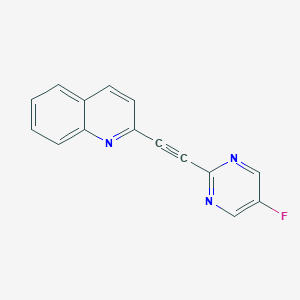 Quinoline,2-[2-(5-fluoro-2-pyrimidinyl)ethynyl]- Quinoline,2-[2-(5-fluoro-2-pyrimidinyl)ethynyl]- | C15H8FN3 | 249.248 | 4 / 0 | 2.7 | Yes |
| 7995 | 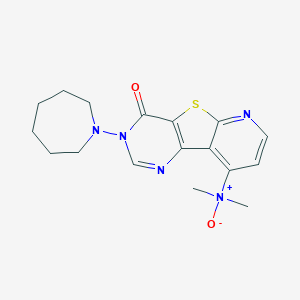 CHEMBL386408 CHEMBL386408 | C17H21N5O2S | 359.448 | 6 / 0 | 2.3 | Yes |
| 8044 | 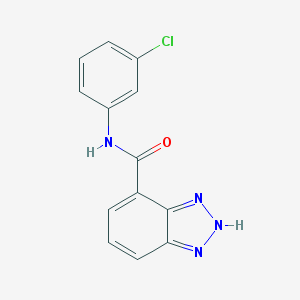 CHEMBL239604 CHEMBL239604 | C13H9ClN4O | 272.692 | 3 / 2 | 2.5 | Yes |
| 8150 | 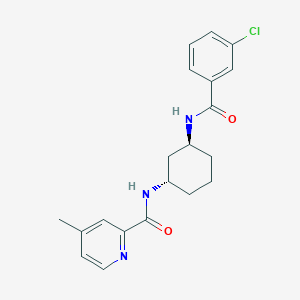 CHEMBL2338601 CHEMBL2338601 | C20H22ClN3O2 | 371.865 | 3 / 2 | 3.8 | Yes |
| 8262 | 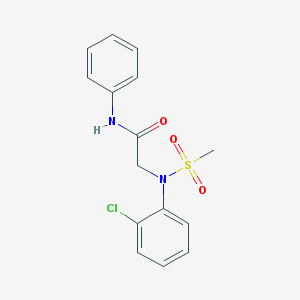 CHEMBL2208408 CHEMBL2208408 | C15H15ClN2O3S | 338.806 | 4 / 1 | 2.6 | Yes |
| 8285 | 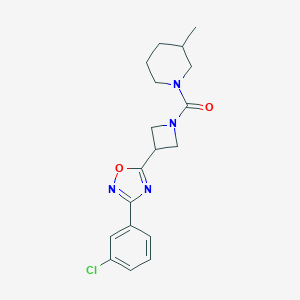 CHEMBL2151622 CHEMBL2151622 | C18H21ClN4O2 | 360.842 | 4 / 0 | 3.1 | Yes |
| 8567 | 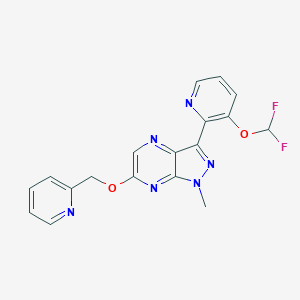 CHEMBL3122214 CHEMBL3122214 | C18H14F2N6O2 | 384.347 | 9 / 0 | 2.2 | Yes |
| 8568 | 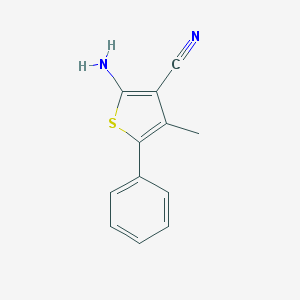 42160-26-9 42160-26-9 | C12H10N2S | 214.286 | 3 / 1 | 3.6 | Yes |
| 536191 | 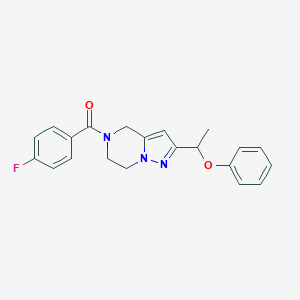 SCHEMBL9926343 SCHEMBL9926343 | C21H20FN3O2 | 365.408 | 4 / 0 | 2.9 | Yes |
| 8694 | 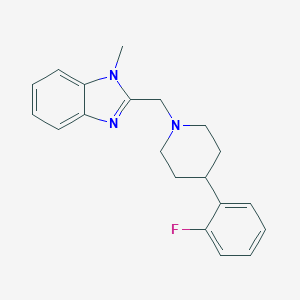 CHEMBL607689 CHEMBL607689 | C20H22FN3 | 323.415 | 3 / 0 | 3.6 | Yes |
| 463932 | 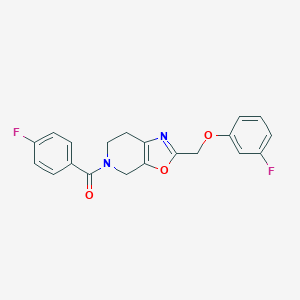 CHEMBL3605277 CHEMBL3605277 | C20H16F2N2O3 | 370.356 | 6 / 0 | 3.1 | Yes |
| 8735 | 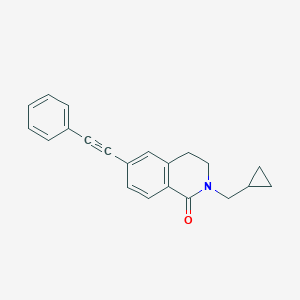 CHEMBL1684105 CHEMBL1684105 | C21H19NO | 301.389 | 1 / 0 | 4.2 | Yes |
| 8746 | 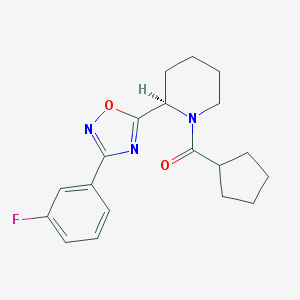 CHEMBL1771682 CHEMBL1771682 | C19H22FN3O2 | 343.402 | 5 / 0 | 3.7 | Yes |
| 8823 | 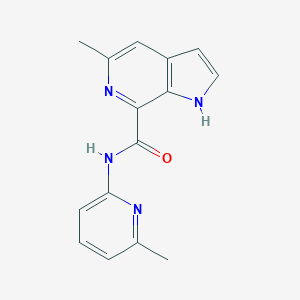 CHEMBL2153782 CHEMBL2153782 | C15H14N4O | 266.304 | 3 / 2 | 2.2 | Yes |
| 463976 | 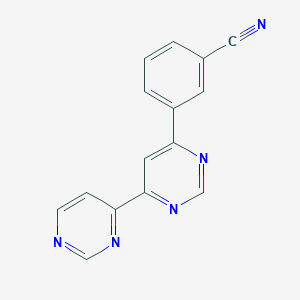 CHEMBL3603928 CHEMBL3603928 | C15H9N5 | 259.272 | 5 / 0 | 1.6 | Yes |
| 464007 | 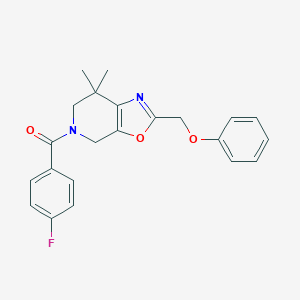 CHEMBL3605300 CHEMBL3605300 | C22H21FN2O3 | 380.419 | 5 / 0 | 3.9 | Yes |
| 9343 | 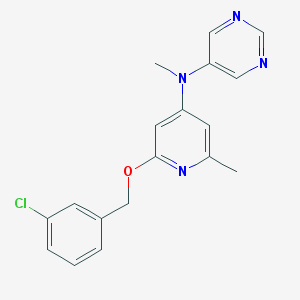 CHEMBL207097 CHEMBL207097 | C18H17ClN4O | 340.811 | 5 / 0 | 3.6 | Yes |
| 9621 | 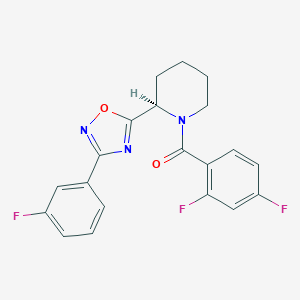 CHEMBL1771692 CHEMBL1771692 | C20H16F3N3O2 | 387.362 | 7 / 0 | 4.0 | Yes |
| 547995 | 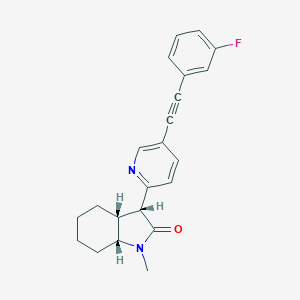 CHEMBL3903257 CHEMBL3903257 | C22H21FN2O | 348.421 | 3 / 0 | 3.9 | Yes |
| 536239 | 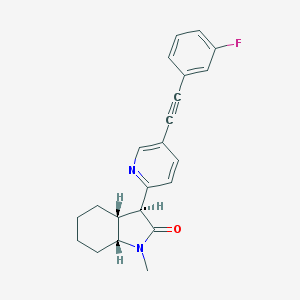 CHEMBL3924488 CHEMBL3924488 | C22H21FN2O | 348.421 | 3 / 0 | 3.9 | Yes |
| 9953 | 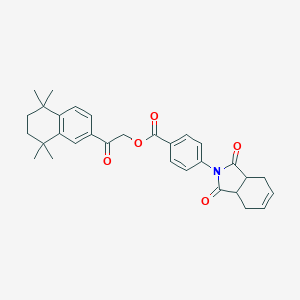 CHEMBL363408 CHEMBL363408 | C31H33NO5 | 499.607 | 5 / 0 | 6.3 | No |
| 9971 | 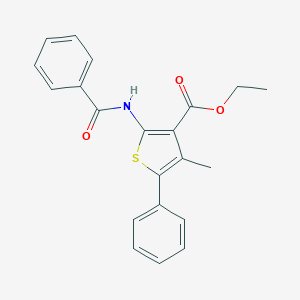 CHEMBL575695 CHEMBL575695 | C21H19NO3S | 365.447 | 4 / 1 | 5.6 | No |
| 10196 | 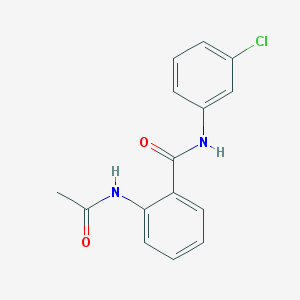 2-acetamido-N-(3-chlorophenyl)benzamide 2-acetamido-N-(3-chlorophenyl)benzamide | C15H13ClN2O2 | 288.731 | 2 / 2 | 3.8 | Yes |
| 10208 | 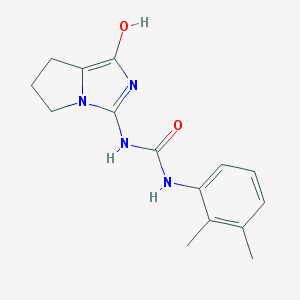 BDBM50179661 BDBM50179661 | C15H18N4O2 | 286.335 | 3 / 3 | 2.0 | Yes |
| 10401 | 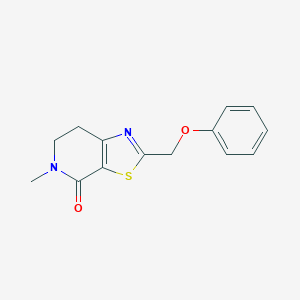 CHEMBL2426616 CHEMBL2426616 | C14H14N2O2S | 274.338 | 4 / 0 | 2.3 | Yes |
| 10610 | 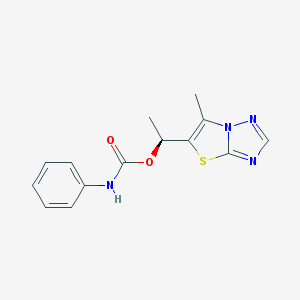 AC1LSLDP AC1LSLDP | C14H14N4O2S | 302.352 | 5 / 1 | 2.8 | Yes |
| 10611 | 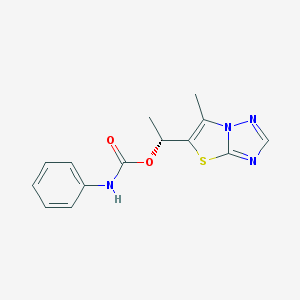 GSK2210875 GSK2210875 | C14H14N4O2S | 302.352 | 5 / 1 | 2.8 | Yes |
| 10613 | 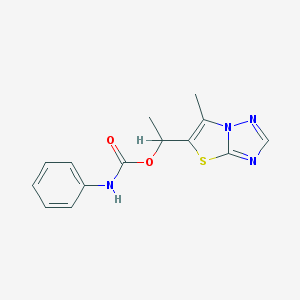 1-(6-methyl[1,3]thiazolo[3,2-b][1,2,4]triazol-5-yl)ethyl N-phenylcarbamate 1-(6-methyl[1,3]thiazolo[3,2-b][1,2,4]triazol-5-yl)ethyl N-phenylcarbamate | C14H14N4O2S | 302.352 | 5 / 1 | 2.8 | Yes |
| 536263 | 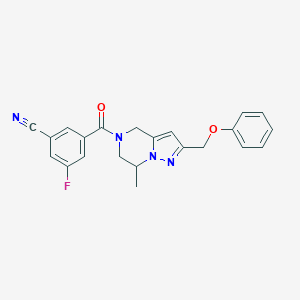 SCHEMBL9926434 SCHEMBL9926434 | C22H19FN4O2 | 390.418 | 5 / 0 | 2.7 | Yes |
| 10817 | 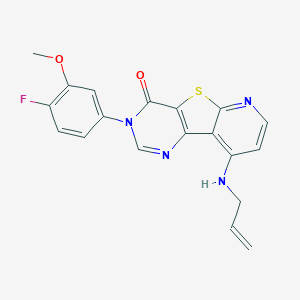 CHEMBL1783862 CHEMBL1783862 | C19H15FN4O2S | 382.413 | 7 / 1 | 3.8 | Yes |
| 11022 | 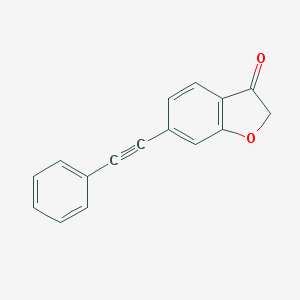 CID 53321026 CID 53321026 | C16H10O2 | 234.254 | 2 / 0 | 3.5 | Yes |
| 464266 | 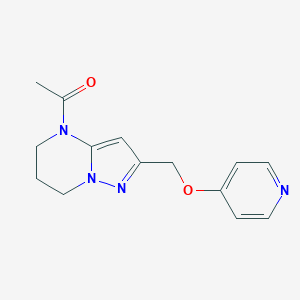 CHEMBL3633967 CHEMBL3633967 | C14H16N4O2 | 272.308 | 4 / 0 | 0.4 | Yes |
| 11689 | 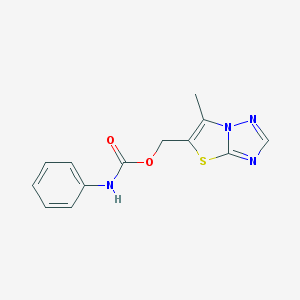 SCHEMBL8030897 SCHEMBL8030897 | C13H12N4O2S | 288.325 | 5 / 1 | 2.4 | Yes |
| 442161 | 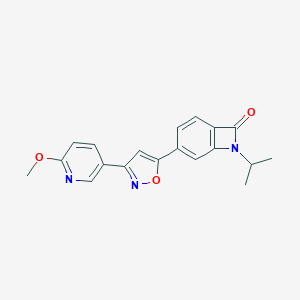 CHEMBL3398289 CHEMBL3398289 | C19H17N3O3 | 335.363 | 5 / 0 | 2.3 | Yes |
| 536309 | 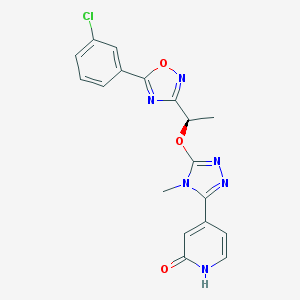 SCHEMBL2959089 SCHEMBL2959089 | C18H15ClN6O3 | 398.807 | 7 / 1 | 1.7 | Yes |
| 12256 | 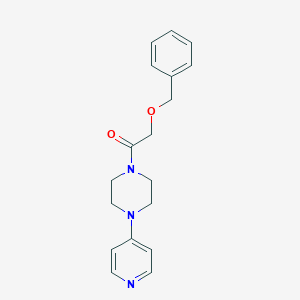 CHEMBL1290788 CHEMBL1290788 | C18H21N3O2 | 311.385 | 4 / 0 | 1.6 | Yes |
| 12551 | 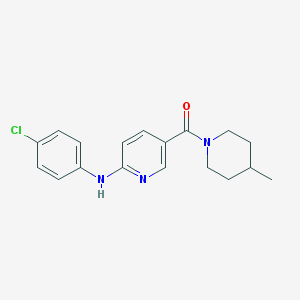 CHEMBL610918 CHEMBL610918 | C18H20ClN3O | 329.828 | 3 / 1 | 4.1 | Yes |
| 464388 | 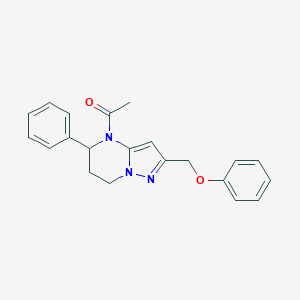 CHEMBL3633978 CHEMBL3633978 | C21H21N3O2 | 347.418 | 3 / 0 | 3.0 | Yes |
| 12906 | 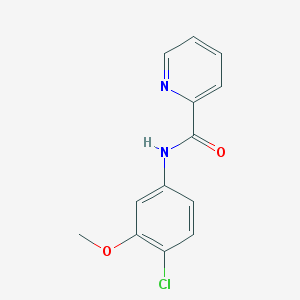 VU 0361737 VU 0361737 | C13H11ClN2O2 | 262.693 | 3 / 1 | 2.6 | Yes |
| 13007 | 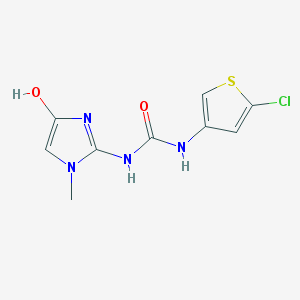 BDBM50201658 BDBM50201658 | C9H9ClN4O2S | 272.707 | 4 / 3 | 1.5 | Yes |
| 13015 | 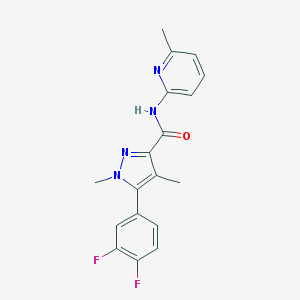 CHEMBL2349537 CHEMBL2349537 | C18H16F2N4O | 342.35 | 5 / 1 | 3.3 | Yes |
| 13082 | 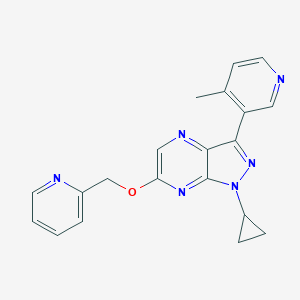 CHEMBL3122219 CHEMBL3122219 | C20H18N6O | 358.405 | 6 / 0 | 2.0 | Yes |
| 13323 | 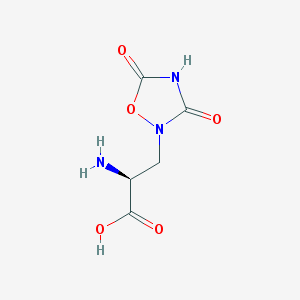 QUISQUALIC ACID QUISQUALIC ACID | C5H7N3O5 | 189.127 | 6 / 3 | -3.9 | Yes |
| 13331 | 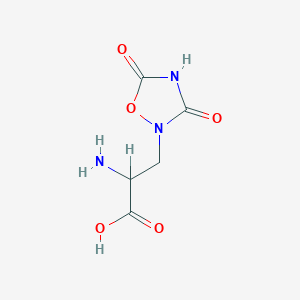 2-amino-3-(3,5-dioxo-1,2,4-oxadiazolidin-2-yl)propanoic acid 2-amino-3-(3,5-dioxo-1,2,4-oxadiazolidin-2-yl)propanoic acid | C5H7N3O5 | 189.127 | 6 / 3 | -3.9 | Yes |
| 13423 | 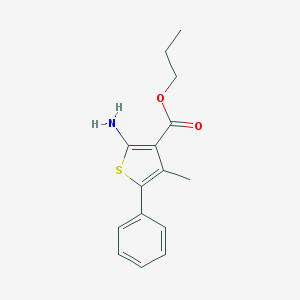 CHEMBL568515 CHEMBL568515 | C15H17NO2S | 275.366 | 4 / 1 | 4.6 | Yes |
| 13460 | 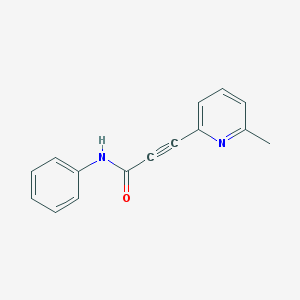 CHEMBL215746 CHEMBL215746 | C15H12N2O | 236.274 | 2 / 1 | 2.7 | Yes |
| 13557 | 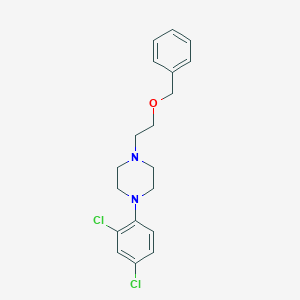 CHEMBL1289784 CHEMBL1289784 | C19H22Cl2N2O | 365.298 | 3 / 0 | 4.5 | Yes |
| 13571 | 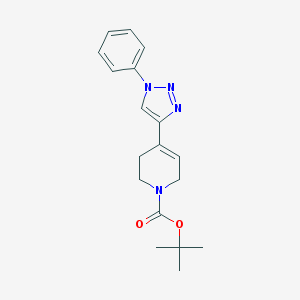 CHEMBL511355 CHEMBL511355 | C18H22N4O2 | 326.4 | 4 / 0 | 2.4 | Yes |
| 13791 | 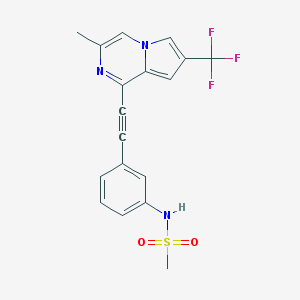 CHEMBL407707 CHEMBL407707 | C18H14F3N3O2S | 393.384 | 7 / 1 | 3.7 | Yes |
| 13879 | 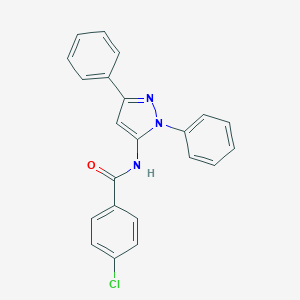 CHEMBL363062 CHEMBL363062 | C22H16ClN3O | 373.84 | 2 / 1 | 5.3 | No |
| 14196 | 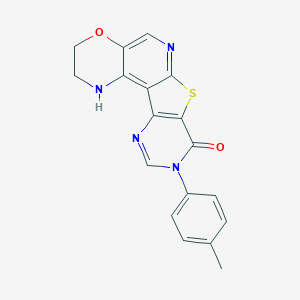 CHEMBL235975 CHEMBL235975 | C18H14N4O2S | 350.396 | 6 / 1 | 3.2 | Yes |
| 14395 | 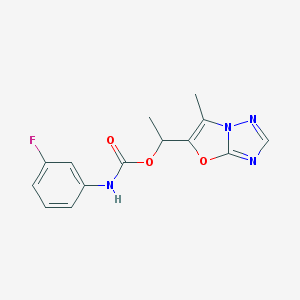 CHEMBL1630064 CHEMBL1630064 | C14H13FN4O3 | 304.281 | 6 / 1 | 2.2 | Yes |
| 14890 | 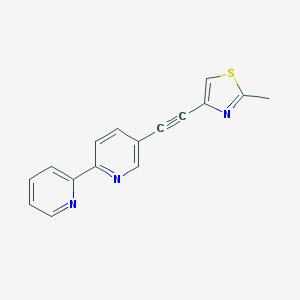 CHEMBL362217 CHEMBL362217 | C16H11N3S | 277.345 | 4 / 0 | 3.0 | Yes |
| 14913 | 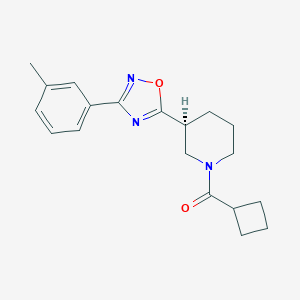 CHEMBL1771621 CHEMBL1771621 | C19H23N3O2 | 325.412 | 4 / 0 | 3.2 | Yes |
| 14914 | 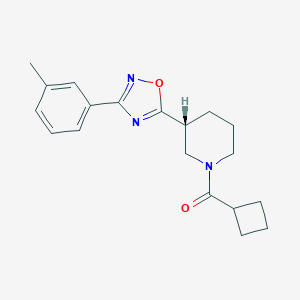 CHEMBL1771622 CHEMBL1771622 | C19H23N3O2 | 325.412 | 4 / 0 | 3.2 | Yes |
| 555541 | 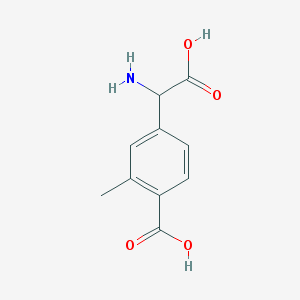 2Me4CPG 2Me4CPG | C10H11NO4 | 209.201 | 5 / 3 | -1.8 | Yes |
| 15055 | 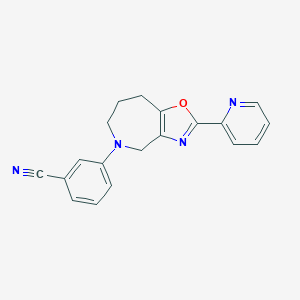 CHEMBL1257504 CHEMBL1257504 | C19H16N4O | 316.364 | 5 / 0 | 2.8 | Yes |
| 15216 | 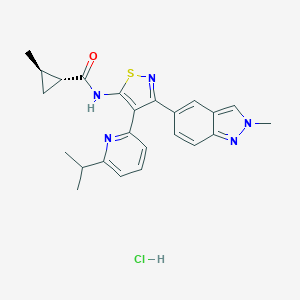 CHEMBL2334981 CHEMBL2334981 | C24H26ClN5OS | 468.016 | 5 / 2 | N/A | N/A |
| 15316 | 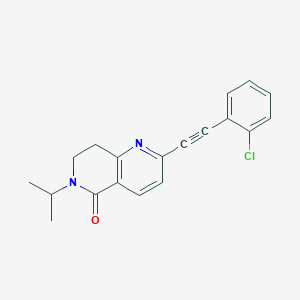 CHEMBL1779862 CHEMBL1779862 | C19H17ClN2O | 324.808 | 2 / 0 | 3.9 | Yes |
| 15349 | 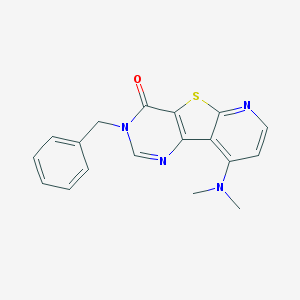 CHEMBL388087 CHEMBL388087 | C18H16N4OS | 336.413 | 5 / 0 | 3.1 | Yes |
| 442253 | 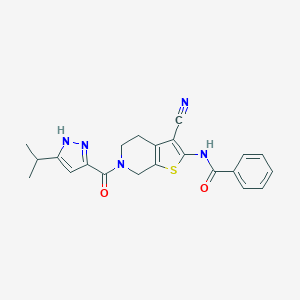 CHEMBL3422865 CHEMBL3422865 | C22H21N5O2S | 419.503 | 5 / 2 | 3.9 | Yes |
zhanglab![]() zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218