

You can:
| Name | D(2) dopamine receptor |
|---|---|
| Species | Rattus norvegicus (Rat) |
| Gene | Drd2 |
| Synonym | D2 receptor D2(415) and D2(444) D2A and D2B D2R Dopamine D2 receptor [ Show all ] |
| Disease | N/A for non-human GPCRs |
| Length | 444 |
| Amino acid sequence | MDPLNLSWYDDDLERQNWSRPFNGSEGKADRPHYNYYAMLLTLLIFIIVFGNVLVCMAVSREKALQTTTNYLIVSLAVADLLVATLVMPWVVYLEVVGEWKFSRIHCDIFVTLDVMMCTASILNLCAISIDRYTAVAMPMLYNTRYSSKRRVTVMIAIVWVLSFTISCPLLFGLNNTDQNECIIANPAFVVYSSIVSFYVPFIVTLLVYIKIYIVLRKRRKRVNTKRSSRAFRANLKTPLKGNCTHPEDMKLCTVIMKSNGSFPVNRRRMDAARRAQELEMEMLSSTSPPERTRYSPIPPSHHQLTLPDPSHHGLHSNPDSPAKPEKNGHAKIVNPRIAKFFEIQTMPNGKTRTSLKTMSRRKLSQQKEKKATQMLAIVLGVFIICWLPFFITHILNIHCDCNIPPVLYSAFTWLGYVNSAVNPIIYTTFNIEFRKAFMKILHC |
| UniProt | P61169 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | N/A |
| 3D structure model | No available structures or models |
| BioLiP | N/A |
| Therapeutic Target Database | N/A |
| ChEMBL | CHEMBL339 |
| IUPHAR | 215 |
| DrugBank | N/A |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 54 | 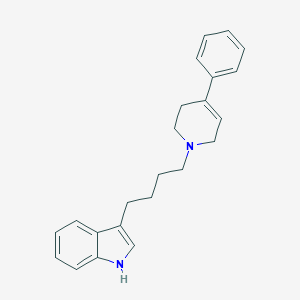 EMD-23448 EMD-23448 | C23H26N2 | 330.475 | 1 / 1 | 4.9 | Yes |
| 60 | 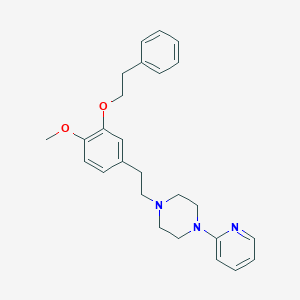 CHEMBL11640 CHEMBL11640 | C26H31N3O2 | 417.553 | 5 / 0 | 5.0 | Yes |
| 84 | 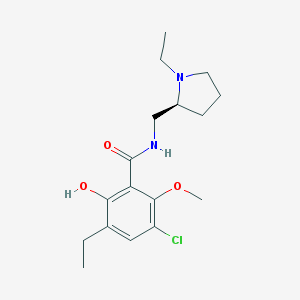 Eticlopride Eticlopride | C17H25ClN2O3 | 340.848 | 4 / 2 | 3.1 | Yes |
| 128 | 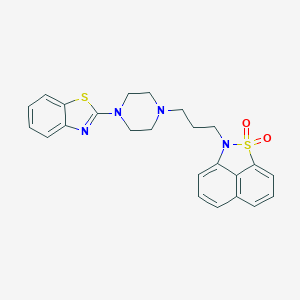 CHEMBL83391 CHEMBL83391 | C24H24N4O2S2 | 464.602 | 7 / 0 | 4.7 | Yes |
| 144 | 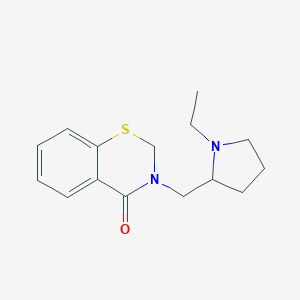 CHEMBL325386 CHEMBL325386 | C15H20N2OS | 276.398 | 3 / 0 | 2.7 | Yes |
| 181 | 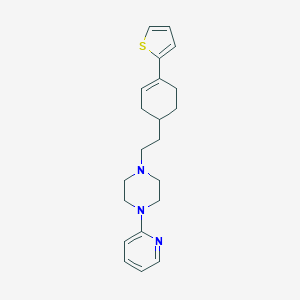 PD-135111 PD-135111 | C21H27N3S | 353.528 | 4 / 0 | 4.3 | Yes |
| 262 | 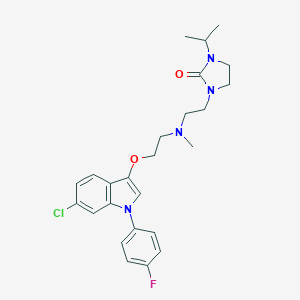 CHEMBL122047 CHEMBL122047 | C25H30ClFN4O2 | 472.989 | 4 / 0 | 4.5 | Yes |
| 272 | 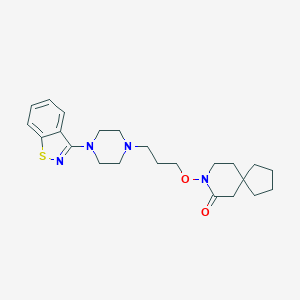 CHEMBL269176 CHEMBL269176 | C23H32N4O2S | 428.595 | 6 / 0 | 4.6 | Yes |
| 274 | 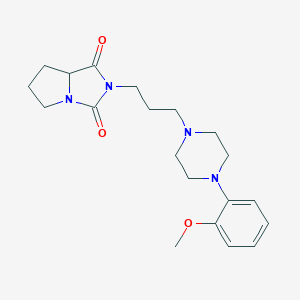 2-{3-[4-(2-methoxyphenyl)-1-piperazinyl]propyl}tetrahydro-1H-pyrrolo[1,2-c]imidazole-1,3(2H)-dione 2-{3-[4-(2-methoxyphenyl)-1-piperazinyl]propyl}tetrahydro-1H-pyrrolo[1,2-c]imidazole-1,3(2H)-dione | C20H28N4O3 | 372.469 | 5 / 0 | 1.8 | Yes |
| 293 | 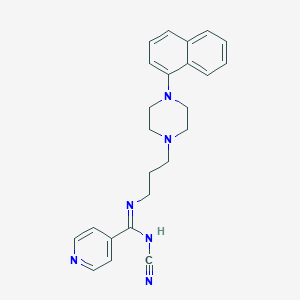 CHEMBL1783352 CHEMBL1783352 | C24H26N6 | 398.514 | 5 / 1 | 3.5 | Yes |
| 332 | 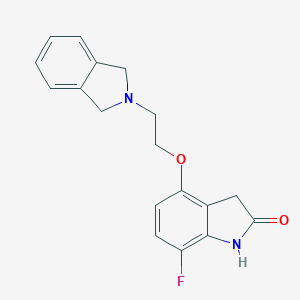 CHEMBL300316 CHEMBL300316 | C18H17FN2O2 | 312.344 | 4 / 1 | 1.9 | Yes |
| 555480 | 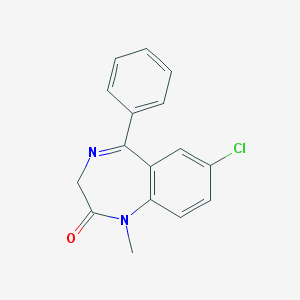 diazepam diazepam | C16H13ClN2O | 284.743 | 2 / 0 | 3.0 | Yes |
| 527 | 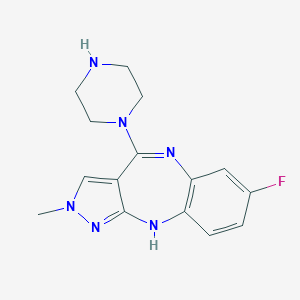 CHEMBL314226 CHEMBL314226 | C15H17FN6 | 300.341 | 5 / 2 | 1.6 | Yes |
| 547906 | 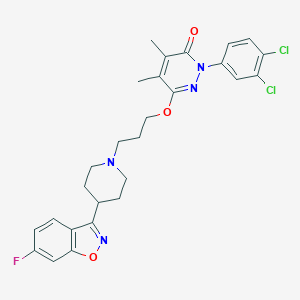 CHEMBL3883388 CHEMBL3883388 | C27H27Cl2FN4O3 | 545.436 | 7 / 0 | 6.0 | No |
| 626 | 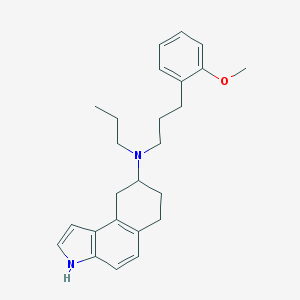 CHEMBL306197 CHEMBL306197 | C25H32N2O | 376.544 | 2 / 1 | 6.0 | No |
| 710 | 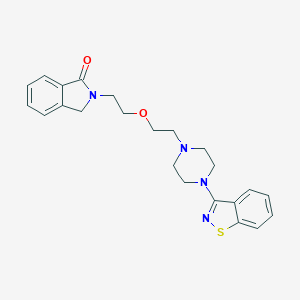 CHEMBL288450 CHEMBL288450 | C23H26N4O2S | 422.547 | 6 / 0 | 3.2 | Yes |
| 744 |  CHEMBL367875 CHEMBL367875 | C21H27N3OS | 369.527 | 4 / 0 | 3.4 | Yes |
| 780 |  CHEMBL542783 CHEMBL542783 | C23H34ClNO3 | 407.979 | 4 / 2 | N/A | N/A |
| 1396 |  CHEMBL37661 CHEMBL37661 | C22H27N | 305.465 | 1 / 0 | 5.5 | No |
| 1398 |  CHEMBL39483 CHEMBL39483 | C22H27N | 305.465 | 1 / 0 | 5.5 | No |
| 1454 |  CHEMBL457025 CHEMBL457025 | C25H35N3O | 393.575 | 4 / 1 | 4.8 | Yes |
| 1456 |  CHEMBL457024 CHEMBL457024 | C25H35N3O | 393.575 | 4 / 1 | 4.8 | Yes |
| 1488 |  CHEMBL3085403 CHEMBL3085403 | C15H22BrClN2O4 | 409.705 | 5 / 4 | N/A | N/A |
| 1492 |  CHEMBL413777 CHEMBL413777 | C23H31NO2 | 353.506 | 3 / 1 | 5.7 | No |
| 1519 | 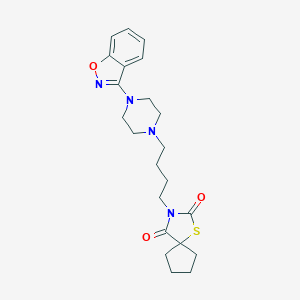 CHEMBL121065 CHEMBL121065 | C22H28N4O3S | 428.551 | 7 / 0 | 3.8 | Yes |
| 1526 | 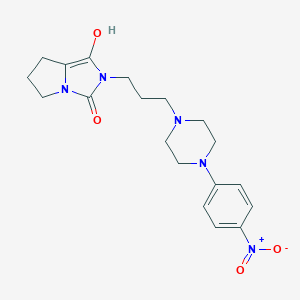 BDBM50054356 BDBM50054356 | C19H25N5O4 | 387.44 | 6 / 1 | 1.6 | Yes |
| 1543 | 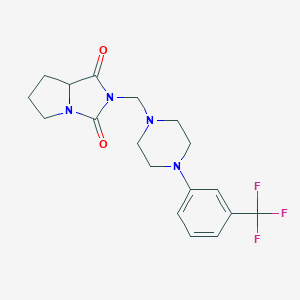 Tetrahydro-2-[[4-[3-(trifluoromethyl)phenyl]piperazin-1-yl]methyl]-1H-pyrrolo[1,2-c]imidazole-1,3(2H)-dione Tetrahydro-2-[[4-[3-(trifluoromethyl)phenyl]piperazin-1-yl]methyl]-1H-pyrrolo[1,2-c]imidazole-1,3(2H)-dione | C18H21F3N4O2 | 382.387 | 7 / 0 | 2.5 | Yes |
| 1644 | 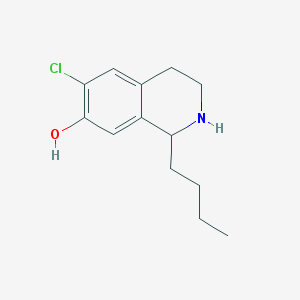 CHEMBL574111 CHEMBL574111 | C13H18ClNO | 239.743 | 2 / 2 | 3.4 | Yes |
| 1812 |  CHEMBL180319 CHEMBL180319 | C15H17N3S2 | 303.442 | 4 / 0 | 3.0 | Yes |
| 521502 |  CHEMBL3741516 CHEMBL3741516 | C27H30FN3O2 | 447.554 | 5 / 0 | 4.7 | Yes |
| 1994 |  CHEMBL545372 CHEMBL545372 | C22H27Cl2N5OS | 480.452 | 6 / 3 | N/A | N/A |
| 2032 |  CHEMBL331611 CHEMBL331611 | C28H28N2 | 392.546 | 1 / 1 | 6.3 | No |
| 2062 |  CHEMBL164195 CHEMBL164195 | C15H18BrN | 292.22 | 1 / 0 | 3.9 | Yes |
| 2104 |  CHEMBL337696 CHEMBL337696 | C20H24N2O7S | 436.479 | 9 / 4 | N/A | N/A |
| 2160 |  CHEMBL81703 CHEMBL81703 | C25H25FN2O2S | 436.545 | 5 / 1 | 3.9 | Yes |
| 2250 |  CHEMBL288244 CHEMBL288244 | C23H22Cl2FN3OS | 478.407 | 5 / 1 | 4.8 | Yes |
| 2354 | 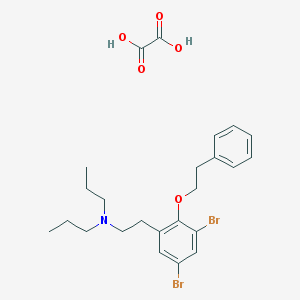 CHEMBL11590 CHEMBL11590 | C24H31Br2NO5 | 573.322 | 6 / 2 | N/A | No |
| 2371 | 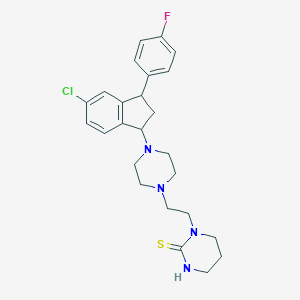 CHEMBL1788128 CHEMBL1788128 | C25H30ClFN4S | 473.051 | 4 / 1 | 4.2 | Yes |
| 2399 | 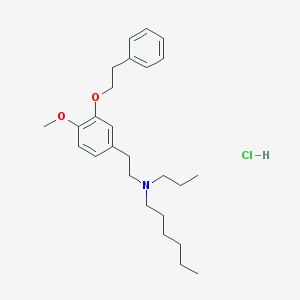 CHEMBL542298 CHEMBL542298 | C26H40ClNO2 | 434.061 | 3 / 1 | N/A | N/A |
| 2947 | 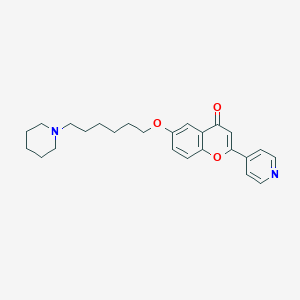 CHEMBL290612 CHEMBL290612 | C25H30N2O3 | 406.526 | 5 / 0 | 4.4 | Yes |
| 2978 | 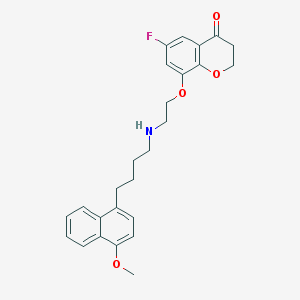 CHEMBL317532 CHEMBL317532 | C26H28FNO4 | 437.511 | 6 / 1 | 4.9 | Yes |
| 3071 | 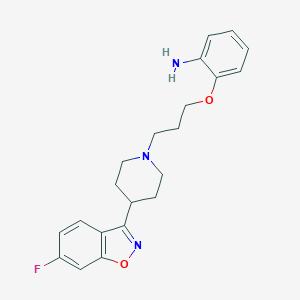 CHEMBL14538 CHEMBL14538 | C21H24FN3O2 | 369.44 | 6 / 1 | 3.8 | Yes |
| 3111 | 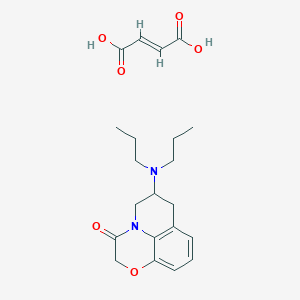 CHEMBL1794795 CHEMBL1794795 | C21H28N2O6 | 404.463 | 7 / 2 | N/A | N/A |
| 3190 | 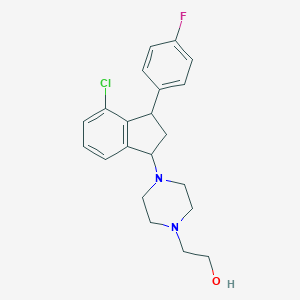 CHEMBL92896 CHEMBL92896 | C21H24ClFN2O | 374.884 | 4 / 1 | 3.5 | Yes |
| 3290 | 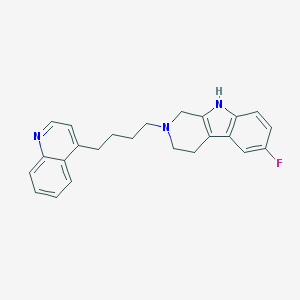 CHEMBL13120 CHEMBL13120 | C24H24FN3 | 373.475 | 3 / 1 | 5.0 | Yes |
| 3416 | 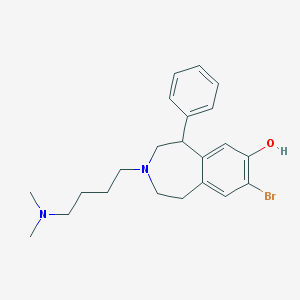 CHEMBL135616 CHEMBL135616 | C22H29BrN2O | 417.391 | 3 / 1 | 4.8 | Yes |
| 3490 | 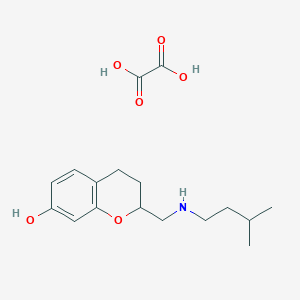 CHEMBL134843 CHEMBL134843 | C17H25NO6 | 339.388 | 7 / 4 | N/A | N/A |
| 555491 | 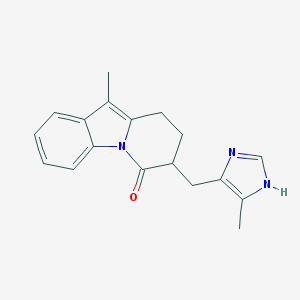 FK 1052 FK 1052 | C18H19N3O | 293.37 | 2 / 1 | 3.2 | Yes |
| 536004 | 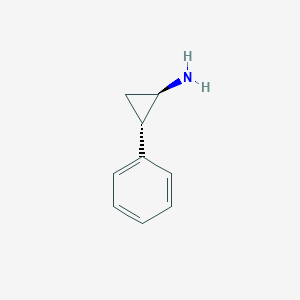 tranylcypromine tranylcypromine | C9H11N | 133.194 | 1 / 1 | 1.5 | Yes |
| 3693 | 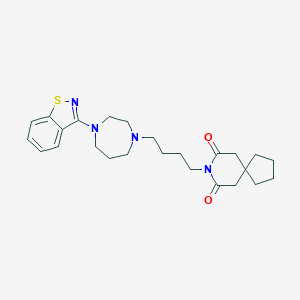 CHEMBL123048 CHEMBL123048 | C25H34N4O2S | 454.633 | 6 / 0 | 4.6 | Yes |
| 3725 | 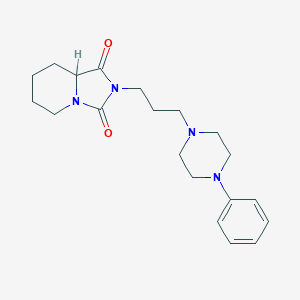 CID 10665976 CID 10665976 | C20H28N4O2 | 356.47 | 4 / 0 | 2.2 | Yes |
| 3752 | 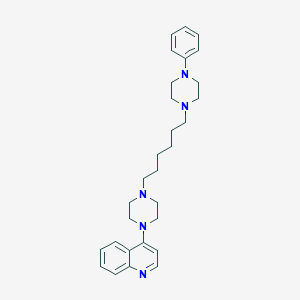 CHEMBL1173017 CHEMBL1173017 | C29H39N5 | 457.666 | 5 / 0 | 5.2 | No |
| 3758 |  CHEMBL542786 CHEMBL542786 | C20H27NO2 | 313.441 | 3 / 1 | 4.5 | Yes |
| 3763 |  CHEMBL566890 CHEMBL566890 | C22H26ClN7O2 | 455.947 | 5 / 0 | 3.0 | Yes |
| 3783 |  CHEMBL1204094 CHEMBL1204094 | C24H29N5O2 | 419.529 | 6 / 0 | 1.4 | Yes |
| 3794 |  CHEMBL60185 CHEMBL60185 | C19H20N2O | 292.382 | 2 / 2 | 3.5 | Yes |
| 3861 | 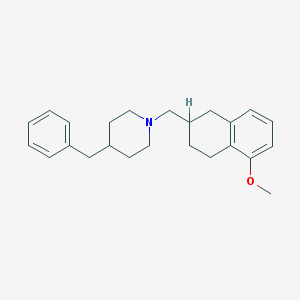 CHEMBL180119 CHEMBL180119 | C24H31NO | 349.518 | 2 / 0 | 5.5 | No |
| 3943 | 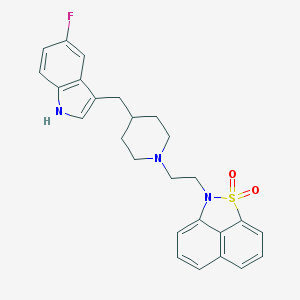 CHEMBL18512 CHEMBL18512 | C26H26FN3O2S | 463.571 | 5 / 1 | 4.9 | Yes |
| 4143 | 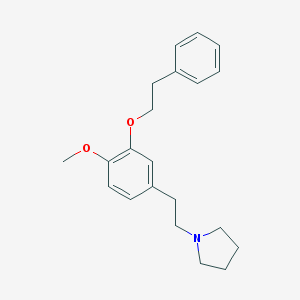 CHEMBL11311 CHEMBL11311 | C21H27NO2 | 325.452 | 3 / 0 | 4.6 | Yes |
| 4232 | 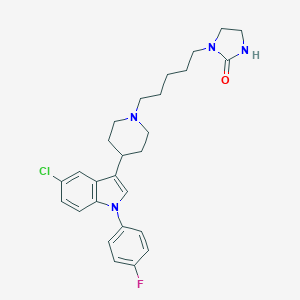 CHEMBL418160 CHEMBL418160 | C27H32ClFN4O | 483.028 | 3 / 1 | 5.1 | No |
| 4236 | 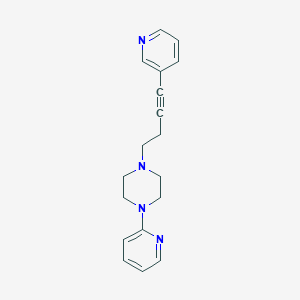 CHEMBL432972 CHEMBL432972 | C18H20N4 | 292.386 | 4 / 0 | 2.4 | Yes |
| 4401 | 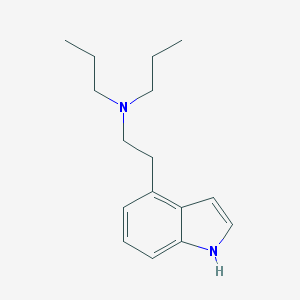 76149-15-0 76149-15-0 | C16H24N2 | 244.382 | 1 / 1 | 4.1 | Yes |
| 4443 | 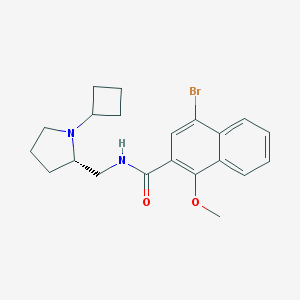 CHEMBL298759 CHEMBL298759 | C21H25BrN2O2 | 417.347 | 3 / 1 | 4.6 | Yes |
| 4444 | 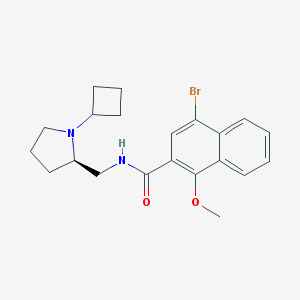 CHEMBL50517 CHEMBL50517 | C21H25BrN2O2 | 417.347 | 3 / 1 | 4.6 | Yes |
| 4509 |  CHEMBL497604 CHEMBL497604 | C24H26N4O2 | 402.498 | 5 / 1 | 4.0 | Yes |
| 4543 |  CHEMBL285712 CHEMBL285712 | C24H25FN2O | 376.475 | 3 / 1 | 4.7 | Yes |
| 441887 |  CHEMBL3392290 CHEMBL3392290 | C27H41N3O6 | 503.64 | 9 / 4 | N/A | No |
| 4599 |  CHEMBL135011 CHEMBL135011 | C16H23NO2 | 261.365 | 3 / 2 | 3.4 | Yes |
| 547947 |  CHEMBL3883429 CHEMBL3883429 | C22H23Cl2N5O2 | 460.359 | 6 / 0 | 3.9 | Yes |
| 4620 |  CHEMBL95112 CHEMBL95112 | C23H27FN2O2S | 414.539 | 6 / 0 | 3.4 | Yes |
| 555500 |  Moperone Moperone | C22H26FNO2 | 355.453 | 4 / 1 | 3.0 | Yes |
| 4783 |  CHEMBL268274 CHEMBL268274 | C17H25N3O | 287.407 | 3 / 1 | 3.0 | Yes |
| 4980 | 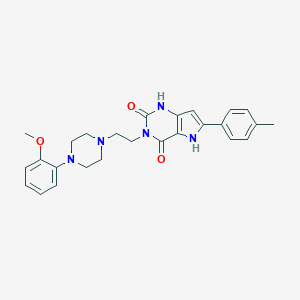 CHEMBL190525 CHEMBL190525 | C26H29N5O3 | 459.55 | 5 / 2 | 3.4 | Yes |
| 5300 | 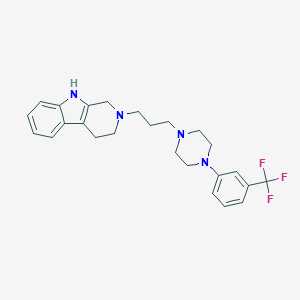 CHEMBL12726 CHEMBL12726 | C25H29F3N4 | 442.53 | 6 / 1 | 4.8 | Yes |
| 5362 | 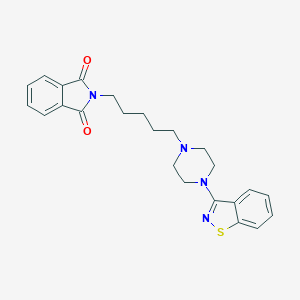 CHEMBL41536 CHEMBL41536 | C24H26N4O2S | 434.558 | 6 / 0 | 4.3 | Yes |
| 5411 | 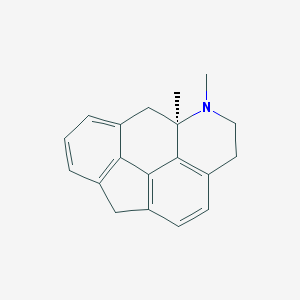 CHEMBL403844 CHEMBL403844 | C19H19N | 261.368 | 1 / 0 | 3.7 | Yes |
| 5435 | 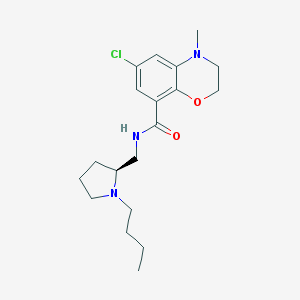 CHEMBL283528 CHEMBL283528 | C19H28ClN3O2 | 365.902 | 4 / 1 | 3.5 | Yes |
| 5438 | 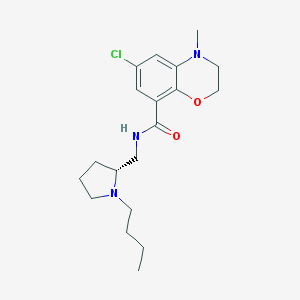 CHEMBL279729 CHEMBL279729 | C19H28ClN3O2 | 365.902 | 4 / 1 | 3.5 | Yes |
| 5453 | 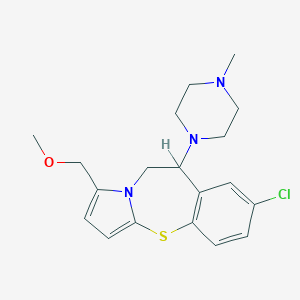 CHEMBL175226 CHEMBL175226 | C19H24ClN3OS | 377.931 | 4 / 0 | 2.8 | Yes |
| 5520 | 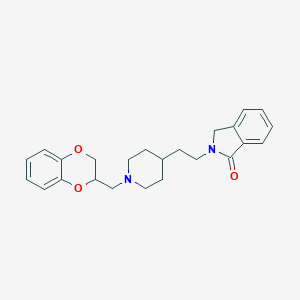 CHEMBL121638 CHEMBL121638 | C24H28N2O3 | 392.499 | 4 / 0 | 3.6 | Yes |
| 5565 | 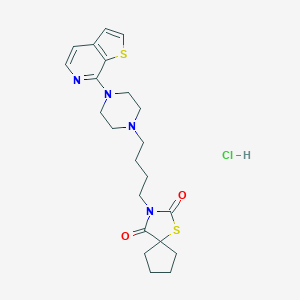 CHEMBL558026 CHEMBL558026 | C22H29ClN4O2S2 | 481.07 | 7 / 1 | N/A | N/A |
| 5698 | 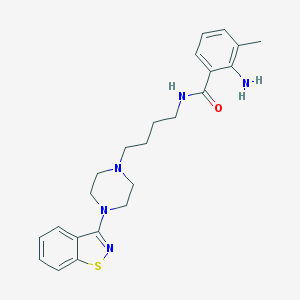 CHEMBL544909 CHEMBL544909 | C23H29N5OS | 423.579 | 6 / 2 | 4.5 | Yes |
| 5709 | 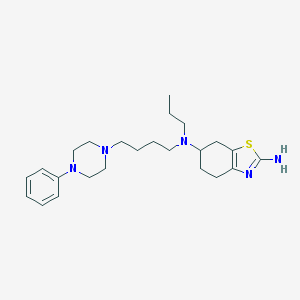 CHEMBL468022 CHEMBL468022 | C24H37N5S | 427.655 | 6 / 1 | 4.7 | Yes |
| 5716 | 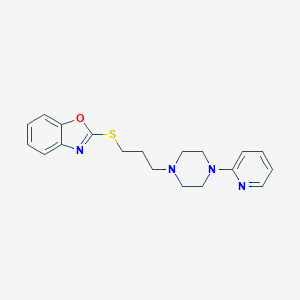 CHEMBL517962 CHEMBL517962 | C19H22N4OS | 354.472 | 6 / 0 | 3.9 | Yes |
| 555506 |  paroxetine paroxetine | C19H20FNO3 | 329.371 | 5 / 1 | 3.5 | Yes |
| 5849 |  CHEMBL93594 CHEMBL93594 | C15H23NO3S | 297.413 | 4 / 0 | 2.9 | Yes |
| 6139 |  CHEMBL3215641 CHEMBL3215641 | C19H24Cl2N2O2S | 415.373 | 5 / 5 | N/A | N/A |
| 6369 |  CHEMBL545036 CHEMBL545036 | C26H28ClNO | 405.966 | 2 / 1 | N/A | N/A |
| 6453 | 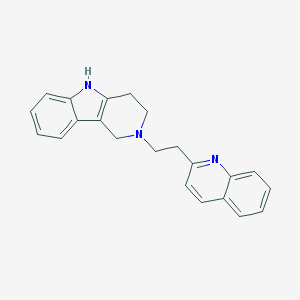 CHEMBL1743950 CHEMBL1743950 | C22H21N3 | 327.431 | 2 / 1 | 4.2 | Yes |
| 6515 | 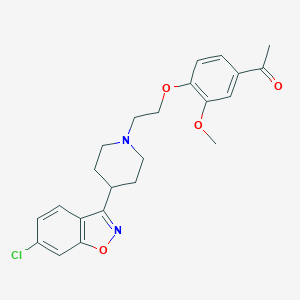 CHEMBL14361 CHEMBL14361 | C23H25ClN2O4 | 428.913 | 6 / 0 | 4.3 | Yes |
| 6879 | 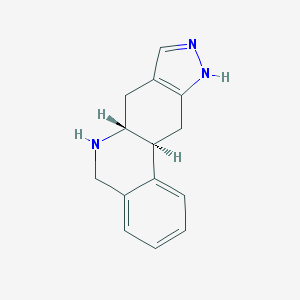 CHEMBL171519 CHEMBL171519 | C14H15N3 | 225.295 | 2 / 2 | 1.5 | Yes |
| 6916 | 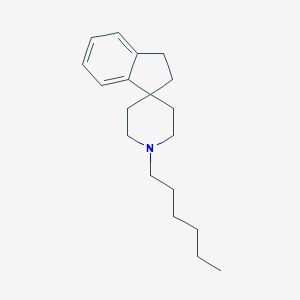 CHEMBL1744015 CHEMBL1744015 | C19H29N | 271.448 | 1 / 0 | 5.4 | No |
| 7245 | 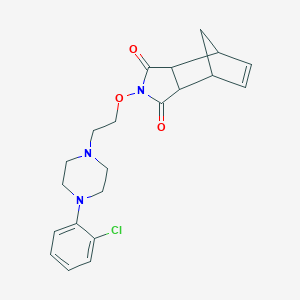 CHEMBL426813 CHEMBL426813 | C21H24ClN3O3 | 401.891 | 5 / 0 | 2.6 | Yes |
| 441992 | 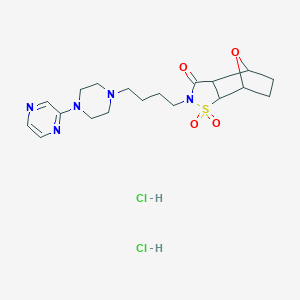 CHEMBL3215745 CHEMBL3215745 | C19H29Cl2N5O4S | 494.432 | 8 / 2 | N/A | N/A |
| 7316 | 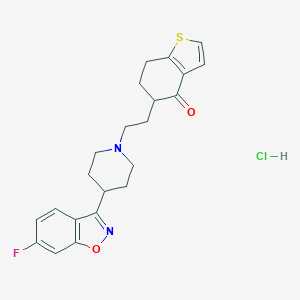 CHEMBL543613 CHEMBL543613 | C22H24ClFN2O2S | 434.954 | 6 / 1 | N/A | N/A |
| 7349 | 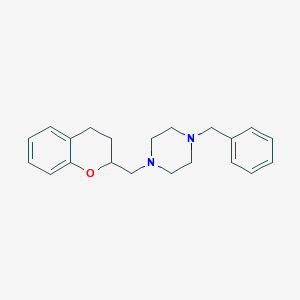 CHEMBL179648 CHEMBL179648 | C21H26N2O | 322.452 | 3 / 0 | 3.7 | Yes |
| 7377 | 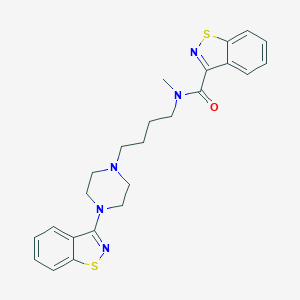 CHEMBL75670 CHEMBL75670 | C24H27N5OS2 | 465.634 | 7 / 0 | 5.2 | No |
| 7407 | 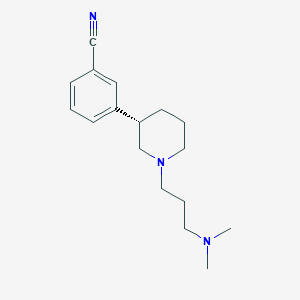 CHEMBL327836 CHEMBL327836 | C17H25N3 | 271.408 | 3 / 0 | 2.6 | Yes |
| 7574 | 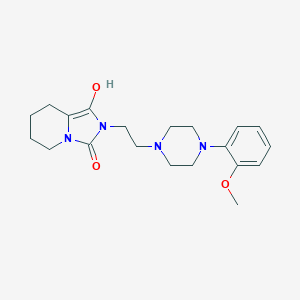 BDBM50054361 BDBM50054361 | C20H28N4O3 | 372.469 | 5 / 1 | 1.7 | Yes |
| 7694 | 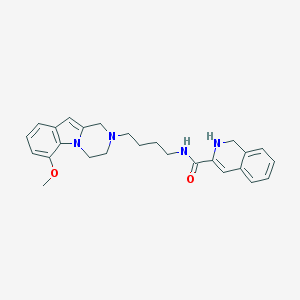 CID 44581146 CID 44581146 | C26H30N4O2 | 430.552 | 4 / 2 | 3.5 | Yes |
zhanglab![]() zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218