

You can:
| Name | P2Y purinoceptor 13 |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | P2RY13 |
| Synonym | G-protein coupled receptor 86 purinergic receptor P2Y, G-protein coupled 13 purinergic receptor P2Y P2Y13 receptor P2Y13 [ Show all ] |
| Disease | N/A |
| Length | 354 |
| Amino acid sequence | MTAAIRRQRELSILPKVTLEAMNTTVMQGFNRSERCPRDTRIVQLVFPALYTVVFLTGILLNTLALWVFVHIPSSSTFIIYLKNTLVADLIMTLMLPFKILSDSHLAPWQLRAFVCRFSSVIFYETMYVGIVLLGLIAFDRFLKIIRPLRNIFLKKPVFAKTVSIFIWFFLFFISLPNTILSNKEATPSSVKKCASLKGPLGLKWHQMVNNICQFIFWTVFILMLVFYVVIAKKVYDSYRKSKSKDRKNNKKLEGKVFVVVAVFFVCFAPFHFARVPYTHSQTNNKTDCRLQNQLFIAKETTLFLAATNICMDPLIYIFLCKKFTEKLPCMQGRKTTASSQENHSSQTDNITLG |
| UniProt | Q9BPV8 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | Q9BPV8 |
| 3D structure model | This predicted structure model is from GPCR-EXP Q9BPV8. |
| BioLiP | N/A |
| Therapeutic Target Database | N/A |
| ChEMBL | N/A |
| IUPHAR | 329 |
| DrugBank | N/A |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 553662 | 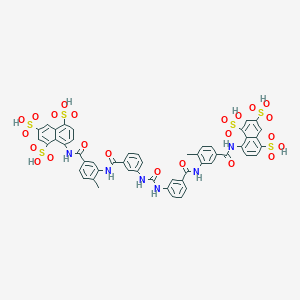 suramin suramin | C51H40N6O23S6 | 1297.26 | 23 / 12 | 1.5 | No |
| 553805 | 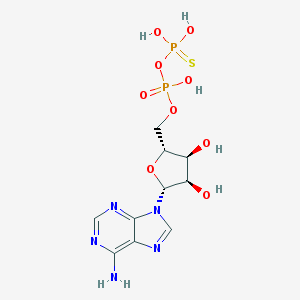 Adpbetas Adpbetas | C10H15N5O9P2S | 443.264 | 14 / 6 | -2.9 | No |
| 554070 | 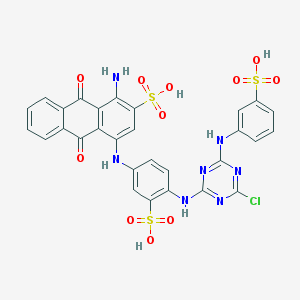 Affi gel blue Affi gel blue | C29H20ClN7O11S3 | 774.147 | 18 / 7 | 4.4 | No |
| 554401 | 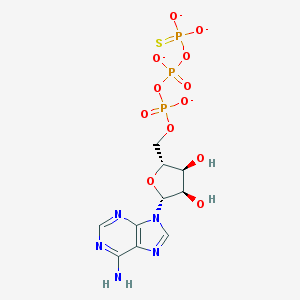 ATPgammaS ATPgammaS | C10H12N5O12P3S-4 | 519.21 | 17 / 3 | -4.3 | No |
| 554530 | 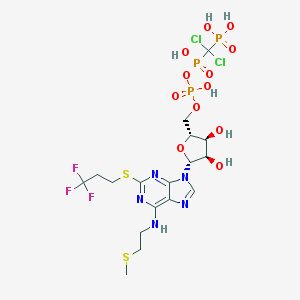 Cangrelor Cangrelor | C17H25Cl2F3N5O12P3S2 | 776.346 | 21 / 7 | -1.0 | No |
| 555165 | 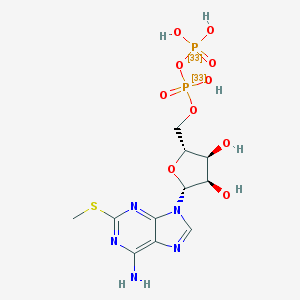 [33P]2-methylthio-ADP [33P]2-methylthio-ADP | C11H17N5O10P2S | 477.285 | 15 / 6 | -3.8 | No |
| 555166 | 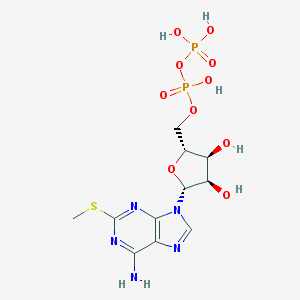 2-MeSADP 2-MeSADP | C11H17N5O10P2S | 473.29 | 15 / 6 | -3.8 | No |
| 555267 | 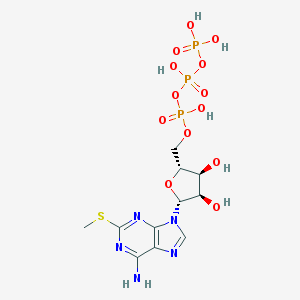 2-Mesatp 2-Mesatp | C11H18N5O13P3S | 553.268 | 18 / 7 | -4.9 | No |
| 555280 | 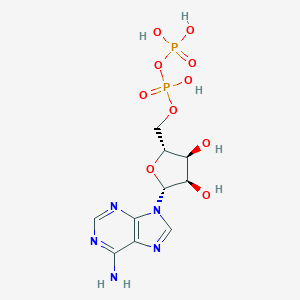 Adenosine 5'-diphosphate Adenosine 5'-diphosphate | C10H15N5O10P2 | 427.203 | 14 / 6 | -4.6 | No |
| 555283 | 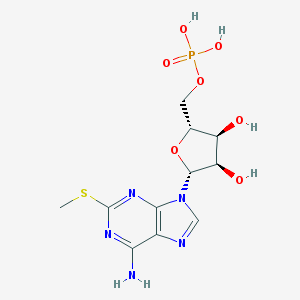 Poly(2'-methylthioadenylic acid) Poly(2'-methylthioadenylic acid) | C11H16N5O7PS | 393.311 | 12 / 5 | -2.7 | No |
| 555350 | 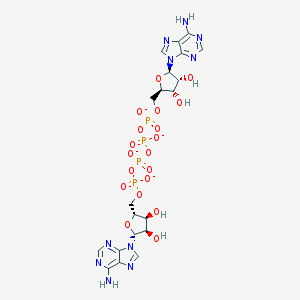 Ap4A Ap4A | C20H24N10O19P4-4 | 832.358 | 27 / 6 | -8.6 | No |
| 555425 | 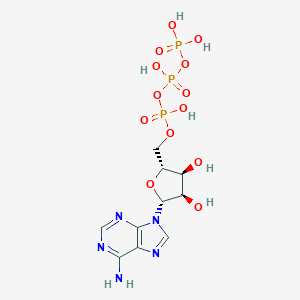 Adenosine triphosphate Adenosine triphosphate | C10H16N5O13P3 | 507.181 | 17 / 7 | -5.7 | No |
zhanglab![]() zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218