

You can:
| Name | SCHEMBL1991690 |
|---|---|
| Molecular formula | C16H16ClN3O |
| IUPAC name | 4-chloro-N-(6-pyrrolidin-3-ylpyridin-3-yl)benzamide |
| Molecular weight | 301.774 |
| Hydrogen bond acceptor | 3 |
| Hydrogen bond donor | 2 |
| XlogP | 2.1 |
| Synonyms | Benzamide, 4-chloro-N-[6-(3-pyrrolidinyl)-3-pyridinyl]- (RS)-4-chloro-N-(6-pyrrolidin-3-yl-pyridin-3-yl)-benzamide ABRIYQXZDGDZAN-UHFFFAOYSA-N 1312561-45-7 US9452980, 140 [ Show all ] |
| Inchi Key | ABRIYQXZDGDZAN-UHFFFAOYSA-N |
| Inchi ID | InChI=1S/C16H16ClN3O/c17-13-3-1-11(2-4-13)16(21)20-14-5-6-15(19-10-14)12-7-8-18-9-12/h1-6,10,12,18H,7-9H2,(H,20,21) |
| PubChem CID | 67239828 |
| ChEMBL | CHEMBL3912684 |
| IUPHAR | N/A |
| BindingDB | 250230 |
| DrugBank | N/A |
Structure | 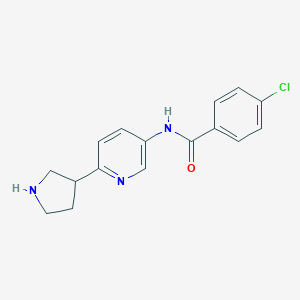 |
| Lipinski's druglikeness | This ligand satisfies Lipinski's rule of five. |
You can:
| GLASS ID | Name | UniProt | Gene | Species | Length |
|---|---|---|---|---|---|
| 535942 | Trace amine-associated receptor 1 | Q923Y9 | Taar1 | Rattus norvegicus (Rat) | 332 |
| 535943 | Trace amine-associated receptor 1 | Q923Y8 | Taar1 | Mus musculus (Mouse) | 332 |
zhanglab![]() zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218