

You can:
| Name | 1-methyl-3-ethylxanthine |
|---|---|
| Molecular formula | C8H10N4O2 |
| IUPAC name | 3-ethyl-1-methyl-7H-purine-2,6-dione |
| Molecular weight | 194.194 |
| Hydrogen bond acceptor | 3 |
| Hydrogen bond donor | 1 |
| XlogP | 0.5 |
| Synonyms | 125573-05-9 CHEMBL334813 3-Ethyl-1-methyl-3,7-dihydro-purine-2,6-dione BDBM50001510 1H-Purine-2,6-dione, 3-ethyl-3,7-dihydro-1-methyl- [ Show all ] |
| Inchi Key | JUZUUEGSTBASHX-UHFFFAOYSA-N |
| Inchi ID | InChI=1S/C8H10N4O2/c1-3-12-6-5(9-4-10-6)7(13)11(2)8(12)14/h4H,3H2,1-2H3,(H,9,10) |
| PubChem CID | 15931309 |
| ChEMBL | CHEMBL334813 |
| IUPHAR | N/A |
| BindingDB | 50001510 |
| DrugBank | N/A |
Structure | 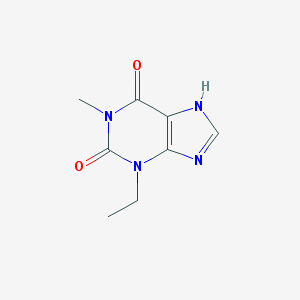 |
| Lipinski's druglikeness | This ligand satisfies Lipinski's rule of five. |
You can:
| GLASS ID | Name | UniProt | Gene | Species | Length |
|---|---|---|---|---|---|
| 160669 | Adenosine receptor A1 | P47745 | ADORA1 | Cavia porcellus (Guinea pig) | 326 |
| 160667 | Adenosine receptor A2a | P29274 | ADORA2A | Homo sapiens (Human) | 412 |
| 160668 | Adenosine receptor A2a | P46616 | ADORA2A | Cavia porcellus (Guinea pig) | 409 |
zhanglab![]() zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218