| Synonyms | Poletoxol
UNII-X7V411JQHV
Polyethylene glycol 2000 octyl phenyl ether
Polyethylene glycol octylphenol ether
Polyoxyethylene (12) octyl phenyl ether
Polyoxyethylene mono(octylphenyl) ether
STL451484
Triton X 165
Triton X-102
Triton,(+)
UNII-7JPC6Y25QS
OP1EO
AN-19902
Peg 4-isooctylphenyl ether
CAS_118-96-7
DSSTox_CID_14085
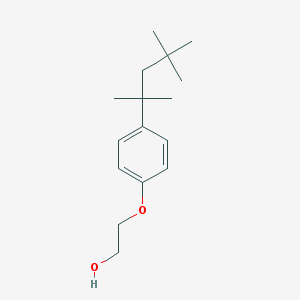
2-(4-(1,1,3,3-Tetramethylbutyl)phenoxy)ethanol
Ethanol, octylphenoxy-
2003274-72-2
Hydrol SW
3,9,12,15,18,21,24,27,30-Decaoxatriacontan-1-ol, 30-[p-(1,1,3,3-tetramethylbutyl)phenyl]-
MCULE-7119685362
4-iso-Octylphenol-mono-ethoxylate 10 microg/mL in Acetone
NSC-5259
48RF3T316O
Octoxynol 1
9002-93-1
Octoxynol-33
AKOS009460920
UNII-GW0EMR6SXY
Poly(oxy-1,2-ethanediyl), alpha-(octylphenyl)-omega-hydroxy-
Polyethylene glycol mono(4-octylphenyl) ether
Polyethylene glycol p-octylphenyl ether
Polyoxyethylene (33) octyl phenyl ether
Preceptin
Tox21_202544
Triton X 405
Triton x-45
TX-017094
PEG-33 Octyl phenyl ether
alpha-(4-(1,1,3,3-Tetramethylbutyl)phenyl)-omega-hydroxypoly(oxy-1,2-ethanediyl)
p-(1,1,3,3-Tetramethylbutyl)phenol ethoxylate
Bio1_000474
PEG-11 Octyl phenyl ether
CHEBI:9750
1017686-87-1
DTXSID1058680
2-[4-(1,1,3,3-tetramethylbutyl)phenoxy]ethan-1-ol
FT-0673247
20CAX7IO75
J-015013
39409-11-5
Neutronyx 605
4-tert-Octylphenol monoethoxylate solution, 1 mug/mL in acetone, analytical standard
Octoxinol (INN)
66057-82-7
Octoxynol 9 [USAN]
9077-65-0
Octoxynols
UNII-QH2U227LZY
Polyethylene glycol (25) octyl phenyl ether
Polyethylene glycol monoether with p-tert-octylphenyl
Polyethyleneglycol 4-(tert-octyl)phenyl ether
Polyoxyethylene (9) octylphenyl ether
SQL994V0M6
Triton X 101
Triton(R) X-114
UNII-480KVF3EBY
Phenol, P-(1,1,3,3-tetramethylbutyl)-, monoether with polyethylene glycol
octylphenoxy polyethoxy ethanol
alpha-(p-(1,1,3,3-Tetramethylbutyl)phenyl)-omega-hydroxypoly(oxyethylene) produced by the condensation of 1 mole of p-(1,1,3,3-tetramethylbutyl)phenol with a range of 30-70 moles of ethylene oxide
PDSP2_001071
C11623
CTK5G8407
13463-07-5
ETHANOL, 2-(P-(1,1,3,3-TETRAMETHYLBUTYL)PHENOXY)-
2-[4-(2,4,4-trimethylpentan-2-yl)phenoxy]ethanol
Glycols, polyethylene, mono(p-(1,1,3,3-tetramethylbutyl)phenyl) ether
26-(Octylphenoxy)-3,6,9,12,15,18,21,24-octaoxahexacosan-1-ol
LS-72946
4-(1,1,3,3-Tetramethylbutyl)phenyl hydroxypoly(oxyethylene)
NSC 5259
4-tert-Octylphenyl peg ether
Octoxinolum [INN-Latin]
Octoxynol-25
AC1L1KOQ
Poly(oxy-1, .alpha.-[4-(1,1,3,3-tetramethylbutyl)phenyl]-.omega.-hydroxy-
X7V411JQHV
Polyethylene glycol 450 octyl phenyl ether
Polyethylene glycol p-(1,3,3-tetramethylbutyl)phenyl ether
Polyoxyethylene (13) octylphenyl ether
Polyoxyethylene mono(octylphenyl) ether (VAN)
Texofor FP 300
Triton X 305
Triton X-305
Triton,(-)
UNII-8419DEW37J
Alfenol 3
OPE 30
Antarox A-200
Peg 4-tert-octylphenyl ether
CAS_5590
DSSTox_GSID_34085
2-(4-tert-Octylphenoxy)ethanol
Ethanol,1,3,3-tetramethylbutyl)phenoxy]-
2055492-07-2
Hyonic pe-250
308104-64-5
NCGC00091012-01
4-tert-Octylphenol Monoethoxylate
NSC5259
66057-68-9
Octoxynol 9 (NF)
9010-42-8
Octoxynol-40
UNII-KI56N6W95G
Poly(oxyethylene)-p-tert-octylphenyl ether
Polyethylene glycol mono(p-(1,1,3,3-tetramethylbutyl)phenyl) ether
Polyethylene glycol p-octylphenyl ether (VAN)
Polyoxyethylene (40) octyl phenyl ether
QH2U227LZY
Triton X
Triton X 45
TRITON(R) X-100
UNII-20CAX7IO75
PEG-40 Octyl phenyl ether
alpha-(P-(1,1,3,3-Tetramethylbutyl)phenoxy)ethanol
p-tert-(Octylphenoxypolyethoxy)ethanol
Bio1_000963
PEG-12 Octyl phenyl ether
CHEMBL39763
111287-03-7
EINECS 264-520-1
2-[4-(1,1,3,3-Tetramethylbutyl)phenoxy]ethanol
FT-0689215
2315-67-5
JYCQQPHGFMYQCF-UHFFFAOYSA-N
3E2NC94VPF
NR7ZWN391G
4-tert-Octylphenol monoethoxylate solution, 10 mug/mL in acetone, analytical standard
Octoxinols
7JPC6Y25QS
Octoxynol-11
935753-04-1
Octyl phenol condensed with l2-l3 moles ethylene oxide
UNII-SQL994V0M6
Polyethylene glycol (33) octyl phenyl ether
Polyethylene glycol mono[p-(1,3,3-tetramethylbutyl)phenyl] ether
Polyoxyethylene (11) octyl phenyl ether
Polyoxyethylene 4-(1,1,3,3-tetramethylbutyl)phenyl ether
ST50406992
Triton X 102
Triton X-100 (TN)
Triton(R) X-405, 70% solution in water
UNII-48RF3T316O
Phenol,1,3,3-tetramethylbutyl)-, monoether with polyethylene glycol
Octylphenoxypolyethoxyethanols
AN-19486
Peg (P-(1,1,3,3-tetramethylbutyl)phenyl) ether
CAS-9002-93-1
D05229
172826-82-3
Ethanol, 2-[4-(1,1,3,3-tetramethylbutyl)phenoxy]-
2-[p-(1,1,3,3-Tetramethylbutyl)phenoxy]ethanol
GW0EMR6SXY
3,6,9,12,15,18,21,24-Octaoxahexacosan-1-ol, 26-(octylphenoxy)-
Marlophen 820
4-iso-Octylphenol-mono-ethoxylate
NSC-406472
480KVF3EBY
Octoxynol
8419DEW37J
Octoxynol-3
AC1Q56RS
Poly(oxy-1,2-ethanediyl), alpha-(4-(1,1,3,3-tetramethylbutyl)phenyl)-omega-hydroxy-
ZINC1532311
Polyethylene glycol 600 octyl phenyl ether
Polyethylene glycol p-1,1,3,3-tetramethylbutylphenyl ether
Polyoxyethylene (25) octyl phenyl ether
Polyoxyethylene octyl phenyl ether
Tox21_111055
Triton X 35
Triton X-405
TX 100
UNII-9T1C662FKS
Alfenol 9
Ortho-gynol
BDBM81480
Peg P-tert-octylphenyl ether
CCRIS 985
DSSTox_RID_79110
2-(p-(1,1,3,3-Tetramethylbutyl)phenoxy)ethanol
Ethoxylated p-tert-octylphenol
2091235-81-1
Igepal CA-630
32-(4-(1,1,3,3-Tetramethylbutyl)phenoxy)-3,6,9,12,15,18,21,24,27,- 30-decaoxadotriacontan-1-ol
NCGC00260093-01
4-tert-Octylphenol monoethoxylate solution
NSC_5590
66057-69-0
Octoxynol 9 [USAN:NF]
9010-43-9
Octoxynol-9
UNII-NR7ZWN391G
Polyethylene glycol (11) octyl phenyl ether
Polyethylene glycol mono(p-tert-octylphenyl) ether
Polyethylene glycol p-tert-octylphenyl ether
Polyoxyethylene (9) octyl phenyl ether
SCHEMBL33822
Triton X 100
Triton X 705
Triton(R) X-100, 98%, for molecular biology, DNAse, RNAse and Protease free
UNII-3E2NC94VPF
PEG-9 Octyl phenyl ether
Octylphenol Ethoxylate
alpha-(P-(1,1,3,3-Tetramethylbutyl)phenyl)-omega-hydroxypoly(oxyethylene)
PDSP1_001087
Bio1_001452
PEG-25 Octyl phenyl ether
Conco nix-100
125692-92-4
Ethanol, 2-(4-(1,1,3,3-tetramethylbutyl)phenoxy)-
2-[4-(2,4,4-Trimethyl-2-pentanyl)phenoxy]ethanol
Glycols, mono[p-(1,1,3,3-tetramethylbutyl)phenyl] ether
26-(4-Octylphenoxy)-3,6,9,12,15,18,21,24-octaoxahexacosan-1-ol
KI56N6W95G
4-(1,1,3,3-Tetramethylbutyl)phenol, ethoxylated
NSC 406472
4-tert-Octylphenyl (2-Hydroxyethyl)ether
Octoxinolum
81398-86-9
Octoxynol-12
9T1C662FKS [ Show all ] |
|---|


![]() zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218