

●I-TASSER ●I-TASSER-MTD ●C-I-TASSER ●CR-I-TASSER ●QUARK ●C-QUARK ●LOMETS ●MUSTER ●CEthreader ●SEGMER ●DeepFold ●DeepFoldRNA ●FoldDesign ●COFACTOR ●COACH ●MetaGO ●TripletGO ●IonCom ●FG-MD ●ModRefiner ●REMO ●DEMO ●DEMO-EM ●SPRING ●COTH ●Threpp ●PEPPI ●BSpred ●ANGLOR ●EDock ●BSP-SLIM ●SAXSTER ●FUpred ●ThreaDom ●ThreaDomEx ●EvoDesign ●BindProf ●BindProfX ●SSIPe ●GPCR-I-TASSER ●MAGELLAN ●ResQ ●STRUM ●DAMpred
●TM-score ●TM-align ●US-align ●MM-align ●RNA-align ●NW-align ●LS-align ●EDTSurf ●MVP ●MVP-Fit ●SPICKER ●HAAD ●PSSpred ●3DRobot ●MR-REX ●I-TASSER-MR ●SVMSEQ ●NeBcon ●ResPRE ●TripletRes ●DeepPotential ●WDL-RF ●ATPbind ●DockRMSD ●DeepMSA ●FASPR ●EM-Refiner ●GPU-I-TASSER
●BioLiP ●E. coli ●GLASS ●GPCR-HGmod ●GPCR-RD ●GPCR-EXP ●Tara-3D ●TM-fold ●DECOYS ●POTENTIAL ●RW/RWplus ●EvoEF ●HPSF ●THE-DB ●ADDRESS ●Alpaca-Antibody ●CASP7 ●CASP8 ●CASP9 ●CASP10 ●CASP11 ●CASP12 ●CASP13 ●CASP14

Updated on: January 7, 2020
GPCR-EXP is a database that specializes in curating experimental and predicted structures of G protein-coupled receptors (GPCR). Structure-related data for experimental structures, such as resolution, publication information, and biological ligand, from the Protein Databank (PDB) are extracted and incorporated into the database. Additionally, all GPCRs from the human genome have been modelled using GPCR-I-TASSER and have been included.
Easy-to-read tables have been constructed to faciliate the browsing of:
Furthermore, database statistics can be accessed to provide a glimpse into the current state of GPCR structural biology, while all PDB structures and data are freely available for download.
November 15, 2019
Apologies for the lack of updates for the past month, as we were undergoing cluster maintenance.
November 6, 2018
UniProt function annotations, including free text annotations and Gene Ontology (GO) terms, are now available for experimentally solved and computationally predicted GPCR structures. GO terms general to all GPCRs are excluded.
August 3, 2018
Binding site predictions from COACH are now available for all predicted GPCR structures.
June 15, 2018
'About' and 'Statistics' pages have been updated.
June 12, 2018
Predicted GPCR structures from the human genome have been added. Accessible from the 'Browse' tab.
June 11, 2018
Layout of database has been completely redesigned again. Each section can be accessed by clicking the tabs.
June 3, 2018
Additional data (structure overlays, ligands, PDB downloads, etc.) accessible by clicking buttons for a popup window on the browse page.
May 12, 2017
GPCR-EXP has been completely redesigned. GPCRs are now browsable in table form.
G protein-coupled receptors (GPCR) are an important superfamily of receptors that are involved in a plethora of physiological functions. Perhaps not surprisingly, they have been implicated in many diseases, such as cancer and diabetes. Having the 3D structure of these receptors plays an enormous role in elucidating its function and medical relevance, as well as facilitating structure-based drug design. In order to address this need, GPCR-EXP database is developed to comprehensively collects and curates data about all known GPCR structures.
GPCR-EXP is designed primarily to be browsed by the user and is organized into two major areas: 1. Experimental PDB structures, and 2. Predicted Structures for the Human Genome. GPCRs are stratified into Class A, Class B, Class C, and Class F for both areas, while predicted structures have the additional categories of 'Other' and 'Putative'. 'Other' GPCRS are experimentally verified but do not belong in the other classes, and 'Putative' GPCRs are those that have not yet been experimentally validated. GPCR structures can be browsed by class. Furthermore, users have the option to browse all structures for either major area all at once in one table.
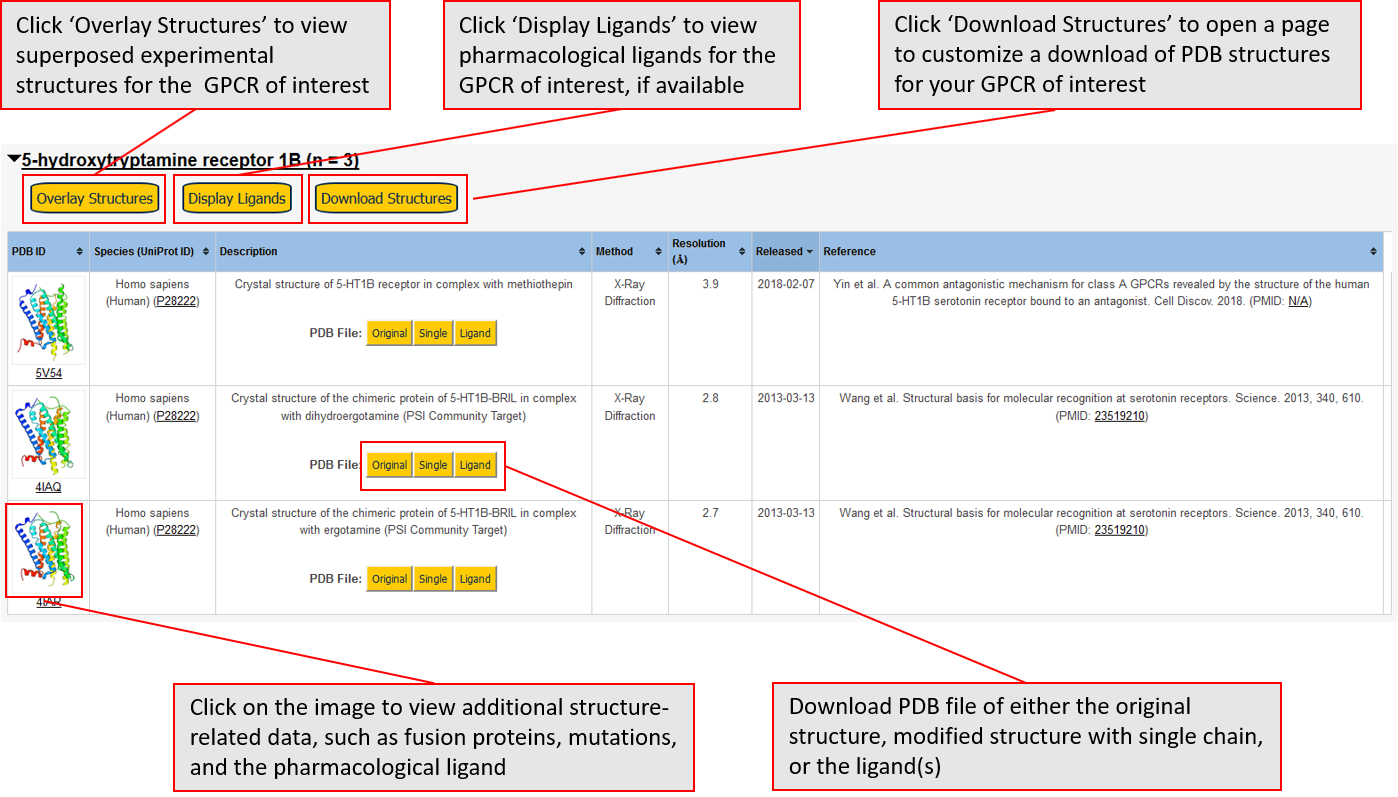
Data for experimental structures are grouped by GPCR if browsing by class. For example, all available PDB structures for the 5-hydroxytryptamine receptor 1B are lumped into the same table. As shown in Figure 1, each PDB structure is represented by one row, where general information is given in each column. The user can view pre-superposed structures by clicking on 'Overlay Structures'. Clicking on 'Display Ligands' will allow the user to view all applicable pharmacological ligands for the structures of the GPCR, whiel clicking on 'Download Structures' provides a customizable download of PDB files. Clicking on the image of the GPCR structure will lead to additional structural and functional data about the PDB structure. Lastly, single PDB files can be downloaded from the table of either original structure, single chain structure, or ligand.
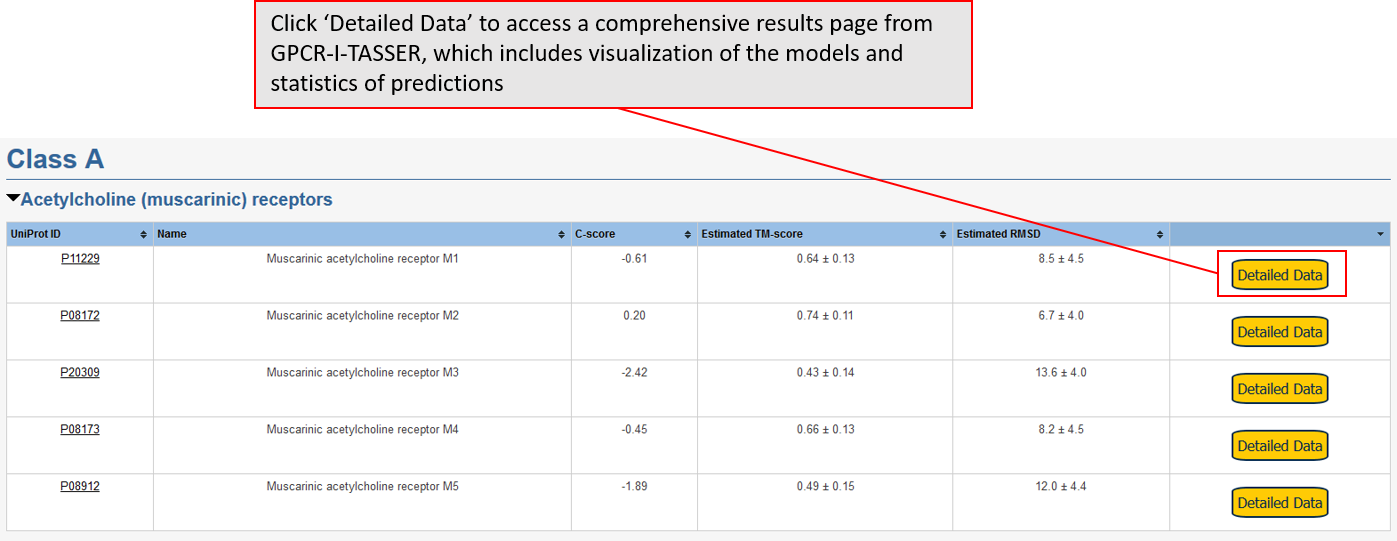
Data for predicted structures are grouped by sub-class for Class A GPCRs but grouped together for the other classes. Figure 2 shows a a grouping of the Class A Acetylcholine receptors, where each row represents a GPCR. General information, such as name and prediction statistics, are given in the columns. The user can click on 'Detailed Data' to access a more comprehensive results page generated by GPCR-I-TASSER.
Wallace Chan, Chengxin Zhang, Mohamed Said, S.M. Mortuza, Yang Zhang. GPCR-EXP: A semi-manually curated database for experimentally-solved and predicted GPCR structures. (2018) In preparation.
Tab-separated values (TSV) file of all extracted structure-related data from PDB
Detailed text file of statistics for experimental GPCR structures from PDB
Unmodified GPCR structures from PDB
GPCR structures modified to contain only a single chain of only the GPCR (no fusion or associated proteins)
GPCRs of the same kind (i.e. mu opioid receptor) are superposed irrespective of species. Includes ligands, as well.
Fragment structures (extracellular domains, incomplete TM domains, etc.) are included for completeness
The top 5 models are included as predicted by GPCR-I-TASSER
| PDB ID | Species (UniProt ID) | Description | Method | Resolution (Å) | Released | Reference | |
|---|---|---|---|---|---|---|---|
 | Homo sapiens (Human) (P28222) | Crystal structure of the chimeric protein of 5-HT1B-BRIL in complex with dihydroergotamine (PSI Community Target) PDB File: | X-Ray Diffraction | 2.8 | 2013-03-13 | Wang et al. Structural basis for molecular recognition at serotonin receptors. Science. 2013, 340, 610. (PMID: 23519210) | |
 | Homo sapiens (Human) (P28222) | Crystal structure of the chimeric protein of 5-HT1B-BRIL in complex with ergotamine (PSI Community Target) PDB File: | X-Ray Diffraction | 2.7 | 2013-03-13 | Wang et al. Structural basis for molecular recognition at serotonin receptors. Science. 2013, 340, 610. (PMID: 23519210) | |
 | Homo sapiens (Human) (P28222) | Crystal structure of 5-HT1B receptor in complex with methiothepin PDB File: | X-Ray Diffraction | 3.9 | 2018-02-07 | Yin et al. A common antagonistic mechanism for class A GPCRs revealed by the structure of the human 5-HT1B serotonin receptor bound to an antagonist. Cell Discov. 2018. (PMID: N/A) | |
 | Homo sapiens (Human) (P28222) | Coupling specificity of heterotrimeric Go to the serotonin 5-HT1B receptor PDB File: | Electron Microscopy | 3.78 | 2018-06-20 | Garcia-Nafria et al. Cryo-EM structure of the serotonin 5-HT1Breceptor coupled to heterotrimeric Go. Nature. 2018, 558, 620. (PMID: 29925951) |
| PDB ID | Species (UniProt ID) | Description | Method | Resolution (Å) | Released | Reference | |
|---|---|---|---|---|---|---|---|
 | Homo sapiens (Human) (P28223) | Crystal structure of 5-HT2AR in complex with risperidone PDB File: | X-Ray Diffraction | 3.0 | 2019-02-13 | Kimura et al. Structures of the 5-HT2Areceptor in complex with the antipsychotics risperidone and zotepine. Nat.Struct.Mol.Biol. 2019, 26, 121. (PMID: 30723326) | |
 | Homo sapiens (Human) (P28223) | Crystal structure of 5-HT2AR in complex with zotepine PDB File: | X-Ray Diffraction | 2.9 | 2019-02-13 | Kimura et al. Structures of the 5-HT2Areceptor in complex with the antipsychotics risperidone and zotepine. Nat.Struct.Mol.Biol. 2019, 26, 121. (PMID: 30723326) |
| PDB ID | Species (UniProt ID) | Description | Method | Resolution (Å) | Released | Reference | |
|---|---|---|---|---|---|---|---|
 | Homo sapiens (Human) (P41595) | Crystal structure of the chimeric protein of 5-HT2B-BRIL in complex with ergotamine PDB File: | X-Ray Diffraction | 2.7 | 2013-03-13 | Wacker et al. Structural features for functional selectivity at serotonin receptors. Science. 2013, 340, 615. (PMID: 23519215) | |
 | Homo sapiens (Human) (P41595) | Crystal structure of the 5-HT2B receptor solved using serial femtosecond crystallography in lipidic cubic phase. PDB File: | X-Ray Diffraction | 2.8 | 2013-12-18 | Liu et al. Serial femtosecond crystallography of G protein-coupled receptors. Science. 2013, 342, 1521. (PMID: 24357322) | |
 | Homo sapiens (Human) (P41595) | Structural Insights into the Extracellular Recognition of the Human Serotonin 2B Receptor by an Antibody PDB File: | X-Ray Diffraction | 3.0 | 2017-07-26 | Ishchenko et al. Structural insights into the extracellular recognition of the human serotonin 2B receptor by an antibody. Proc. Natl. Acad. Sci. U.S.A. 2017, 114, 8223. (PMID: 28716900) | |
 | Homo sapiens (Human) (P41595) | Crystal structure of the LSD-bound 5-HT2B receptor PDB File: | X-Ray Diffraction | 2.9 | 2017-02-01 | Wacker et al. Crystal Structure of an LSD-Bound Human Serotonin Receptor. Cell. 2017, 168, 377. (PMID: 28129538) | |
 | Homo sapiens (Human) (P41595) | Structural Determinants of Activation and Biased Agonism at the 5-HT2B Receptor PDB File: | X-Ray Diffraction | 3.1 | 2018-08-29 | McCorvy et al. Structural determinants of 5-HT2Breceptor activation and biased agonism. Nat. Struct. Mol. Biol. 2018. (PMID: 30127358) | |
 | Homo sapiens (Human) (P41595) | Structural Determinants of Activation and Biased Agonism at the 5-HT2B Receptor PDB File: | X-Ray Diffraction | 2.92 | 2018-08-29 | McCorvy et al. Structural determinants of 5-HT2Breceptor activation and biased agonism. Nat. Struct. Mol. Biol. 2018. (PMID: 30127358) | |
 | Homo sapiens (Human) (P41595) | Structural Determinants of Activation and Biased Agonism at the 5-HT2B Receptor PDB File: | X-Ray Diffraction | 3.1 | 2018-08-29 | McCorvy et al. Structural determinants of 5-HT2Breceptor activation and biased agonism. Nat. Struct. Mol. Biol. 2018. (PMID: 30127358) | |
 | Homo sapiens (Human) (P41595) | Structural Determinants of Activation and Biased Agonism at the 5-HT2B Receptor PDB File: | X-Ray Diffraction | 3.19 | 2018-08-29 | McCorvy et al. Structural determinants of 5-HT2Breceptor activation and biased agonism. Nat. Struct. Mol. Biol. 2018. (PMID: 30127358) |
| PDB ID | Species (UniProt ID) | Description | Method | Resolution (Å) | Released | Reference | |
|---|---|---|---|---|---|---|---|
 | Homo sapiens (Human) (P28335) | Crystal structure of 5-HT2C in complex with ergotamine PDB File: | X-Ray Diffraction | 3.0 | 2018-02-14 | Peng et al. 5-HT2C Receptor Structures Reveal the Structural Basis of GPCR Polypharmacology. Cell. 2018, 172, 719. (PMID: 29398112) | |
 | Homo sapiens (Human) (P28335) | Crystal structure of 5-HT2C in complex with ritanserin PDB File: | X-Ray Diffraction | 2.7 | 2018-02-14 | Peng et al. 5-HT2C Receptor Structures Reveal the Structural Basis of GPCR Polypharmacology. Cell. 2018, 172, 719. (PMID: 29398112) |
| PDB ID | Species (UniProt ID) | Description | Method | Resolution (Å) | Released | Reference | |
|---|---|---|---|---|---|---|---|
 | Homo sapiens (Human) (P30542) | Crystal structure of stabilized A1 receptor in complex with PSB36 at 3.3A resolution PDB File: | X-Ray Diffraction | 3.3 | 2017-07-26 | Cheng et al. Structures of Human A1 and A2A Adenosine Receptors with Xanthines Reveal Determinants of Selectivity. Structure. 2017, 25, 1275. (PMID: 28712806) | |
 | Homo sapiens (Human) (P30542) | Crystal structure of the human adenosine A1 receptor A1AR-bRIL in complex with the covalent antagonist DU172 at 3.2A resolution PDB File: | X-Ray Diffraction | 3.2 | 2017-03-01 | Glukhova et al. Structure of the Adenosine A1 Receptor Reveals the Basis for Subtype Selectivity. Cell. 2017, 168, 867. (PMID: 28235198) | |
 | Homo sapiens (Human) (P30542) | Cryo-EM structure of the human adenosine A1 receptor-Gi2-protein complex bound to its endogenous agonist PDB File: | Electron Microscopy | 3.6 | 2018-06-20 | Draper-Joyce et al. Structure of the adenosine-bound human adenosine A1receptor-Gicomplex. Nature. 2018. (PMID: 29925945) |
| PDB ID | Species (UniProt ID) | Description | Method | Resolution (Å) | Released | Reference | |
|---|---|---|---|---|---|---|---|
 | Homo sapiens (Human) (P29274) | Thermostabilised HUMAN A2a Receptor with adenosine bound PDB File: | X-Ray Diffraction | 3.0 | 2011-05-18 | Lebon et al. Agonist-Bound Adenosine A(2A) Receptor Structures Reveal Common Features of Gpcr Activation. Nature. 2011, 474, 521. (PMID: 21593763) | |
 | Homo sapiens (Human) (P29274) | Thermostabilised HUMAN A2a Receptor with NECA bound PDB File: | X-Ray Diffraction | 2.6 | 2011-05-18 | Lebon et al. Agonist-Bound Adenosine A(2A) Receptor Structures Reveal Common Features of Gpcr Activation. Nature. 2011, 474, 521. (PMID: 21593763) | |
 | Homo sapiens (Human) (P29274) | The 2.6 A Crystal Structure of a Human A2A Adenosine Receptor bound to ZM241385. PDB File: | X-Ray Diffraction | 2.6 | 2008-10-14 | Jaakola et al. The 26 Angstrom Crystal Structure of a Human A2A Adenosine Receptor Bound to an Antagonist. Science. 2008, 322, 1211. (PMID: 18832607) | |
 | Homo sapiens (Human) (P29274) | Thermostabilised Adenosine A2A Receptor PDB File: | X-Ray Diffraction | 3.3 | 2011-09-07 | Dore et al. Structure of the adenosine A(2A) receptor in complex with ZM241385 and the xanthines XAC and caffeine. Structure. 2011, 19, 1283. (PMID: 21885291) | |
 | Homo sapiens (Human) (P29274) | Agonist bound structure of the human adenosine A2a receptor PDB File: | X-Ray Diffraction | 2.71 | 2011-03-09 | Xu et al. Structure of an agonist-bound human A2A adenosine receptor. Science. 2011, 332, 322. (PMID: 21393508) | |
 | Homo sapiens (Human) (P29274) | Thermostabilised adenosine A2A receptor in complex with XAC PDB File: | X-Ray Diffraction | 3.31 | 2011-09-07 | Dore et al. Structure of the adenosine A(2A) receptor in complex with ZM241385 and the xanthines XAC and caffeine. Structure. 2011, 19, 1283. (PMID: 21885291) | |
 | Homo sapiens (Human) (P29274) | Thermostabilised adenosine A2A receptor in complex with caffeine PDB File: | X-Ray Diffraction | 3.6 | 2011-09-07 | Dore et al. Structure of the adenosine A(2A) receptor in complex with ZM241385 and the xanthines XAC and caffeine. Structure. 2011, 19, 1283. (PMID: 21885291) | |
 | Homo sapiens (Human) (P29274) | Thermostabilised Adenosine A2A receptor in complex with 6-(2,6-Dimethylpyridin-4-yl)-5-phenyl-1,2,4-triazin-3-amine PDB File: | X-Ray Diffraction | 3.27 | 2012-03-21 | Congreve et al. Discovery of 1,2,4-Triazine Derivatives as Adenosine A(2A) Antagonists using Structure Based Drug Design. J.Med.Chem. 2012, 55, 1898. (PMID: 22220592) | |
 | Homo sapiens (Human) (P29274) | Thermostabilised Adenosine A2A receptor in complex with 4-(3-amino-5-phenyl-1,2,4-triazin-6-yl)-2-chlorophenol PDB File: | X-Ray Diffraction | 3.34 | 2012-03-21 | Congreve et al. Discovery of 1,2,4-Triazine Derivatives as Adenosine A(2A) Antagonists using Structure Based Drug Design. J.Med.Chem. 2012, 55, 1898. (PMID: 22220592) | |
 | Homo sapiens (Human) (P29274) | Crystal structure of human adenosine A2A receptor with an allosteric inverse-agonist antibody at 2.7 A resolution PDB File: | X-Ray Diffraction | 2.7 | 2012-02-01 | Hino et al. G-protein-coupled receptor inactivation by an allosteric inverse-agonist antibody. Nature. 2012, 482, 237. (PMID: 22286059) | |
 | Homo sapiens (Human) (P29274) | Crystal structure of human adenosine A2A receptor with an allosteric inverse-agonist antibody at 3.1 A resolution PDB File: | X-Ray Diffraction | 3.1 | 2012-02-01 | Hino et al. G-protein-coupled receptor inactivation by an allosteric inverse-agonist antibody. Nature. 2012, 482, 237. (PMID: 22286059) | |
 | Homo sapiens (Human) (P29274) | Crystal structure of the chimeric protein of A2aAR-BRIL in complex with ZM241385 at 1.8A resolution PDB File: | X-Ray Diffraction | 1.8 | 2012-07-25 | Liu et al. Structural basis for allosteric regulation of GPCRs by sodium ions. Science. 2012, 337, 232. (PMID: 22798613) | |
 | Homo sapiens (Human) (P29274) | Thermostabilised HUMAN A2a Receptor with CGS21680 bound PDB File: | X-Ray Diffraction | 2.6 | 2015-04-08 | Lebon et al. Molecular Determinants of Cgs21680 Binding to the Human Adenosine A2A Receptor. Mol.Pharmacol. 2015, 87, 907. (PMID: 25762024) | |
 | Homo sapiens (Human) (P29274) | Thermostabilised HUMAN A2a Receptor with CGS21680 bound PDB File: | X-Ray Diffraction | 2.6 | 2015-04-08 | Lebon et al. Molecular Determinants of Cgs21680 Binding to the Human Adenosine A2A Receptor. Mol.Pharmacol. 2015, 87, 907. (PMID: 25762024) | |
 | Homo sapiens (Human) (P29274) | Structure of the adenosine A2A receptor bound to an engineered G protein PDB File: | X-Ray Diffraction | 3.4 | 2016-08-03 | Carpenter et al. Structure of the Adenosine A2A Receptor Bound to an Engineered G Protein. Nature. 2016, 536, 104. (PMID: 27462812) | |
 | Homo sapiens (Human) (P29274) | Crystal structure of stabilized A2A adenosine receptor A2AR-StaR2-bRIL in complex with ZM241385 at 1.7A resolution PDB File: | X-Ray Diffraction | 1.72 | 2016-06-29 | Segala et al. Controlling the Dissociation of Ligands from the Adenosine A2A Receptor through Modulation of Salt Bridge Strength. J.Med.Chem. 2016, 59, 6470. (PMID: 27312113) | |
 | Homo sapiens (Human) (P29274) | Crystal structure of stabilized A2A adenosine receptor A2AR-StaR2-bRIL in complex with compound 12c at 1.9A resolution PDB File: | X-Ray Diffraction | 1.9 | 2016-06-29 | Segala et al. Controlling the Dissociation of Ligands from the Adenosine A2A Receptor through Modulation of Salt Bridge Strength. J.Med.Chem. 2016, 59, 6470. (PMID: 27312113) | |
 | Homo sapiens (Human) (P29274) | Crystal structure of stabilized A2A adenosine receptor A2AR-StaR2-bRIL in complex with compound 12f at 2.0A resolution PDB File: | X-Ray Diffraction | 2.0 | 2016-06-29 | Segala et al. Controlling the Dissociation of Ligands from the Adenosine A2A Receptor through Modulation of Salt Bridge Strength. J.Med.Chem. 2016, 59, 6470. (PMID: 27312113) | |
 | Homo sapiens (Human) (P29274) | Crystal structure of stabilized A2A adenosine receptor A2AR-StaR2-bRIL in complex with compound 12b at 2.2A resolution PDB File: | X-Ray Diffraction | 2.2 | 2016-06-29 | Segala et al. Controlling the Dissociation of Ligands from the Adenosine A2A Receptor through Modulation of Salt Bridge Strength. J.Med.Chem. 2016, 59, 6470. (PMID: 27312113) | |
 | Homo sapiens (Human) (P29274) | Crystal structure of stabilized A2A adenosine receptor A2AR-StaR2-bRIL in complex with compound 12x at 2.1A resolution PDB File: | X-Ray Diffraction | 2.1 | 2016-06-29 | Segala et al. Controlling the Dissociation of Ligands from the Adenosine A2A Receptor through Modulation of Salt Bridge Strength. J.Med.Chem. 2016, 59, 6470. (PMID: 27312113) | |
 | Homo sapiens (Human) (P29274) | Crystal structure of the chimeric protein of A2aAR-BRIL with bound iodide ions PDB File: | X-Ray Diffraction | 2.8 | 2017-05-31 | Melnikov et al. Fast iodide-SAD phasing for high-throughput membrane protein structure determination. Sci Adv. 2017, 3, e1602952. (PMID: 28508075) | |
 | Homo sapiens (Human) (P29274) | 2.5 angstrom A2a adenosine receptor structure with sulfur SAD phasing using XFEL data PDB File: | X-Ray Diffraction | 2.5 | 2016-09-21 | Batyuk et al. Native phasing of x-ray free-electron laser data for a G protein-coupled receptor. Sci Adv. 2016, 2, e1600292. (PMID: 27679816) | |
 | Homo sapiens (Human) (P29274) | 2.5 angstrom A2a adenosine receptor structure with MR phasing using XFEL data PDB File: | X-Ray Diffraction | 2.5 | 2016-09-21 | Batyuk et al. Native phasing of x-ray free-electron laser data for a G protein-coupled receptor. Sci Adv. 2016, 2, e1600292. (PMID: 27679816) | |
 | Homo sapiens (Human) (P29274) | 1.9 angstrom A2a adenosine receptor structure with sulfur SAD phasing and phase extension using XFEL data PDB File: | X-Ray Diffraction | 1.9 | 2016-09-21 | Batyuk et al. Native phasing of x-ray free-electron laser data for a G protein-coupled receptor. Sci Adv. 2016, 2, e1600292. (PMID: 27679816) | |
 | Homo sapiens (Human) (P29274) | 1.9A angstrom A2a adenosine receptor structure with MR phasing using XFEL data PDB File: | X-Ray Diffraction | 1.9 | 2016-09-21 | Batyuk et al. Native phasing of x-ray free-electron laser data for a G protein-coupled receptor. Sci Adv. 2016, 2, e1600292. (PMID: 27679816) | |
 | Homo sapiens (Human) (P29274) | Crystal structure of stabilized A2A adenosine receptor A2AR-StaR2-bRIL in complex with theophylline at 2.0A resolution PDB File: | X-Ray Diffraction | 2.0 | 2017-07-26 | Cheng et al. Structures of Human A1 and A2A Adenosine Receptors with Xanthines Reveal Determinants of Selectivity. Structure. 2017, 25, 1275. (PMID: 28712806) | |
 | Homo sapiens (Human) (P29274) | Crystal structure of stabilized A2A adenosine receptor A2AR-StaR2-bRIL in complex with caffeine at 2.1A resolution PDB File: | X-Ray Diffraction | 2.1 | 2017-07-26 | Cheng et al. Structures of Human A1 and A2A Adenosine Receptors with Xanthines Reveal Determinants of Selectivity. Structure. 2017, 25, 1275. (PMID: 28712806) | |
 | Homo sapiens (Human) (P29274) | Crystal structure of stabilized A2A adenosine receptor A2AR-StaR2-bRIL in complex with PSB36 at 2.8A resolution PDB File: | X-Ray Diffraction | 2.8 | 2017-07-26 | Cheng et al. Structures of Human A1 and A2A Adenosine Receptors with Xanthines Reveal Determinants of Selectivity. Structure. 2017, 25, 1275. (PMID: 28712806) | |
 | Homo sapiens (Human) (P29274) | A2A Adenosine receptor room-temperature structure determined by serial millisecond crystallography PDB File: | X-Ray Diffraction | 2.14 | 2017-09-27 | Weinert et al. Serial millisecond crystallography for routine room-temperature structure determination at synchrotrons. Nat Commun. 2017, 8, 542. (PMID: 28912485) | |
 | Homo sapiens (Human) (P29274) | A2A Adenosine receptor cryo structure PDB File: | X-Ray Diffraction | 1.95 | 2017-09-27 | Weinert et al. Serial millisecond crystallography for routine room-temperature structure determination at synchrotrons. Nat Commun. 2017, 8, 542. (PMID: 28912485) | |
 | Homo sapiens (Human) (P29274) | A2A Adenosine receptor room-temperature structure determined by serial femtosecond crystallography PDB File: | X-Ray Diffraction | 1.7 | 2017-09-27 | Weinert et al. Serial millisecond crystallography for routine room-temperature structure determination at synchrotrons. Nat Commun. 2017, 8, 542. (PMID: 28912485) | |
 | Homo sapiens (Human) (P29274) | Structure of the A2A-StaR2-bRIL562-ZM241385 complex at 1.86A obtained from in meso soaking experiments. PDB File: | X-Ray Diffraction | 1.87 | 2018-01-17 | Rucktooa et al. Towards high throughput GPCR crystallography: In Meso soaking of Adenosine A2A Receptor crystals. Sci Rep. 2018, 8, 41. (PMID: 29311713) | |
 | Homo sapiens (Human) (P29274) | Structure of the A2A-StaR2-bRIL562-Vipadenant complex at 2.6A obtained from in meso soaking experiments. PDB File: | X-Ray Diffraction | 2.6 | 2018-01-17 | Rucktooa et al. Towards high throughput GPCR crystallography: In Meso soaking of Adenosine A2A Receptor crystals. Sci Rep. 2018, 8, 41. (PMID: 29311713) | |
 | Homo sapiens (Human) (P29274) | Structure of the A2A-StaR2-bRIL562-Tozadenant complex at 3.1A obtained from in meso soaking experiments. PDB File: | X-Ray Diffraction | 3.1 | 2018-01-17 | Rucktooa et al. Towards high throughput GPCR crystallography: In Meso soaking of Adenosine A2A Receptor crystals. Sci Rep. 2018, 8, 41. (PMID: 29311713) | |
 | Homo sapiens (Human) (P29274) | Structure of the A2A-StaR2-bRIL562-LUAA47070 complex at 2.0A obtained from in meso soaking experiments. PDB File: | X-Ray Diffraction | 2.0 | 2018-01-17 | Rucktooa et al. Towards high throughput GPCR crystallography: In Meso soaking of Adenosine A2A Receptor crystals. Sci Rep. 2018, 8, 41. (PMID: 29311713) | |
 | Homo sapiens (Human) (P29274) | Structure of the A2A-StaR2-bRIL562-Compound 4e complex at 1.9A obtained from bespoke co-crystallisation experiments. PDB File: | X-Ray Diffraction | 1.9 | 2018-01-17 | Rucktooa et al. Towards high throughput GPCR crystallography: In Meso soaking of Adenosine A2A Receptor crystals. Sci Rep. 2018, 8, 41. (PMID: 29311713) | |
 | Homo sapiens (Human) (P29274) | Structure of the A2A-StaR2-bRIL562-Compound 4e complex at 2.1A obtained from in meso soaking experiments (1 hour soak). PDB File: | X-Ray Diffraction | 2.1 | 2018-01-17 | Rucktooa et al. Towards high throughput GPCR crystallography: In Meso soaking of Adenosine A2A Receptor crystals. Sci Rep. 2018, 8, 41. (PMID: 29311713) | |
 | Homo sapiens (Human) (P29274) | Structure of the A2A-StaR2-bRIL562-Compound 4e complex at 1.86A obtained from in meso soaking experiments (24 hour soak). PDB File: | X-Ray Diffraction | 2.0 | 2018-01-17 | Rucktooa et al. Towards high throughput GPCR crystallography: In Meso soaking of Adenosine A2A Receptor crystals. Sci Rep. 2018, 8, 41. (PMID: 29311713) | |
 | Homo sapiens (Human) (P29274) | Crystal structure of adenosine A2A receptor bound to a novel triazole-carboximidamide antagonist PDB File: | X-Ray Diffraction | 3.5 | 2017-02-08 | Sun et al. Crystal structure of the adenosine A2A receptor bound to an antagonist reveals a potential allosteric pocket. Proc. Natl. Acad. Sci. U.S.A. 2017, 114, 2066. (PMID: 28167788) | |
 | Homo sapiens (Human) (P29274) | Serial Millisecond Crystallography of Membrane and Soluble Protein Micro-crystals using Synchrotron Radiation PDB File: | X-Ray Diffraction | 3.2 | 2017-05-24 | Martin-Garcia et al. Serial millisecond crystallography of membrane and soluble protein microcrystals using synchrotron radiation. IUCrJ. 2017, 4, 439. (PMID: 28875031) | |
 | Homo sapiens (Human) (P29274) | 2.35-Angstrom In situ Mylar structure of human A2A adenosine receptor at 100 K PDB File: | X-Ray Diffraction | 2.35 | 2017-12-13 | Broecker et al. High-throughput in situ X-ray screening of and data collection from protein crystals at room temperature and under cryogenic conditions. Nat Protoc. 2018, 13, 260. (PMID: 29300389) | |
 | Homo sapiens (Human) (P29274) | Agonist bound human A2a adenosine receptor with D52N mutation at 2.60 A resolution PDB File: | X-Ray Diffraction | 2.6 | 2018-02-21 | White et al. Structural Connection between Activation Microswitch and Allosteric Sodium Site in GPCR Signaling. Structure. 2018, 26, 259. (PMID: 29395784) | |
 | Homo sapiens (Human) (P29274) | Agonist bound human A2a adenosine receptor with S91A mutation at 2.90 A resolution PDB File: | X-Ray Diffraction | 2.9 | 2018-02-21 | White et al. Structural Connection between Activation Microswitch and Allosteric Sodium Site in GPCR Signaling. Structure. 2018, 26, 259. (PMID: 29395784) | |
 | Homo sapiens (Human) (P29274) | Crystal structure of A2AAR-BRIL in complex with the antagonist ZM241385 produced from Pichia pastoris PDB File: | X-Ray Diffraction | 2.51 | 2018-01-10 | Eddy et al. Allosteric Coupling of Drug Binding and Intracellular Signaling in the A2A Adenosine Receptor. Cell. 2018, 172, 68. (PMID: 29290469) | |
 | Homo sapiens (Human) (P29274) | Cryo-EM structure of the adenosine A2A receptor bound to a miniGs heterotrimer PDB File: | Electron Microscopy | 4.11 | 2018-05-16 | Garcia-Nafria et al. Cryo-EM structure of the adenosine A2Areceptor coupled to an engineered heterotrimeric G protein. Elife. 2018, 7, . (PMID: 29726815) | |
 | Homo sapiens (Human) (P29274) | Crystal Structure of the A2A-StaR2-bRIL562 in complex with AZD4635 at 2.0A resolution PDB File: | X-Ray Diffraction | 2.0 | 2019-06-26 | Borodovsky et al. Preclinical pharmacodynamics and antitumor activity of AZD4635, a novel adenosine 2A receptor inhibitor that reverses adenosine mediated immune suppression. To Be Published. (PMID: N/A) | |
 | Homo sapiens (Human) (P29274) | Structure of human A2A adenosine receptor in complex with ZM241385 obtained from SFX experiments under atmospheric pressure PDB File: | X-Ray Diffraction | 2.25 | 2019-10-30 | Shimazu et al. High-viscosity sample-injection device for serial femtosecond crystallography at atmospheric pressure. J.Appl.Crystallogr. 2019, 52, 1280. (PMID: 31798359) | |
 | Homo sapiens (Human) (P29274) | High-viscosity injector-based Pink Beam Serial Crystallography of Micro-crystals at a Synchrotron Radiation Source PDB File: | X-Ray Diffraction | 4.2 | 2019-04-24 | Martin-Garcia et al. High-viscosity injector-based pink-beam serial crystallography of microcrystals at a synchrotron radiation source. Iucrj. 2019, 6, 412. (PMID: 31098022) | |
 | Homo sapiens (Human) (P29274) | XFEL A2aR structure by ligand exchange from LUF5843 to ZM241385. PDB File: | X-Ray Diffraction | 1.85 | 2019-11-13 | Ishchenko et al. Toward G protein-coupled receptor structure-based drug design using X-ray lasers. Iucrj. 2019, 6, 1106. (PMID: 31709066) |
| PDB ID | Species (UniProt ID) | Description | Method | Resolution (Å) | Released | Reference | |
|---|---|---|---|---|---|---|---|
 | Homo sapiens (Human) (P35414) | Structure of apelin receptor in complex with agonist peptide PDB File: | X-Ray Diffraction | 2.6 | 2017-05-31 | Ma et al. Structural Basis for Apelin Control of the Human Apelin Receptor. Structure. 2017, 25, 858. (PMID: 28528775) |
| PDB ID | Species (UniProt ID) | Description | Method | Resolution (Å) | Released | Reference | |
|---|---|---|---|---|---|---|---|
 | Meleagris gallopavo (Wild turkey) (P07700) | TURKEY BETA1 ADRENERGIC RECEPTOR WITH STABILISING MUTATIONS AND BOUND CYANOPINDOLOL PDB File: | X-Ray Diffraction | 2.7 | 2008-06-24 | Warne et al. Structure of a Beta1-Adrenergic G-Protein-Coupled Receptor. Nature. 2008, 454, 486. (PMID: 18594507) | |
 | Meleagris gallopavo (Wild turkey) (P07700) | TURKEY BETA1 ADRENERGIC RECEPTOR WITH STABILISING MUTATIONS AND BOUND PARTIAL AGONIST DOBUTAMINE (CRYSTAL DOB92) PDB File: | X-Ray Diffraction | 2.5 | 2011-01-12 | Warne et al. The Structural Basis for Agonist and Partial Agonist Action on a Beta1-Adrenergic Receptor. Nature. 2011, 469, 241. (PMID: 21228877) | |
 | Meleagris gallopavo (Wild turkey) (P07700) | TURKEY BETA1 ADRENERGIC RECEPTOR WITH STABILISING MUTATIONS AND BOUND PARTIAL AGONIST DOBUTAMINE (CRYSTAL DOB102) PDB File: | X-Ray Diffraction | 2.6 | 2011-03-30 | Warne et al. The Structural Basis for Agonist and Partial Agonist Action on a Beta1-Adrenergic Receptor. Nature. 2011, 469, 241. (PMID: 21228877) | |
 | Meleagris gallopavo (Wild turkey) (P07700) | TURKEY BETA1 ADRENERGIC RECEPTOR WITH STABILISING MUTATIONS AND BOUND AGONIST CARMOTEROL PDB File: | X-Ray Diffraction | 2.6 | 2011-01-12 | Warne et al. The Structural Basis for Agonist and Partial Agonist Action on a Beta1-Adrenergic Receptor. Nature. 2011, 469, 241. (PMID: 21228877) | |
 | Meleagris gallopavo (Wild turkey) (P07700) | TURKEY BETA1 ADRENERGIC RECEPTOR WITH STABILISING MUTATIONS AND BOUND AGONIST ISOPRENALINE PDB File: | X-Ray Diffraction | 2.85 | 2011-01-12 | Warne et al. The Structural Basis for Agonist and Partial Agonist Action on a Beta1-Adrenergic Receptor. Nature. 2011, 469, 241. (PMID: 21228877) | |
 | Meleagris gallopavo (Wild turkey) (P07700) | TURKEY BETA1 ADRENERGIC RECEPTOR WITH STABILISING MUTATIONS AND BOUND PARTIAL AGONIST SALBUTAMOL PDB File: | X-Ray Diffraction | 3.05 | 2011-01-12 | Warne et al. The Structural Basis for Agonist and Partial Agonist Action on a Beta1-Adrenergic Receptor. Nature. 2011, 469, 241. (PMID: 21228877) | |
 | Meleagris gallopavo (Wild turkey) (P07700) | TURKEY BETA1 ADRENERGIC RECEPTOR WITH STABILISING MUTATIONS AND BOUND ANTAGONIST CARAZOLOL PDB File: | X-Ray Diffraction | 3.0 | 2011-06-01 | Moukhametzianov et al. Two Distinct Conformations of Helix 6 Observed in Antagonist-Bound Structures of a Beta-1- Adrenergic Receptor. Proc.Natl.Acad.Sci.USA. 2011, 108, 8228. (PMID: 21540331) | |
 | Meleagris gallopavo (Wild turkey) (P07700) | TURKEY BETA1 ADRENERGIC RECEPTOR WITH STABILISING MUTATIONS AND BOUND ANTAGONIST CYANOPINDOLOL PDB File: | X-Ray Diffraction | 3.25 | 2011-06-01 | Moukhametzianov et al. Two Distinct Conformations of Helix 6 Observed in Antagonist-Bound Structures of a Beta-1- Adrenergic Receptor. Proc.Natl.Acad.Sci.USA. 2011, 108, 8228. (PMID: 21540331) | |
 | Meleagris gallopavo (Wild turkey) (P07700) | TURKEY BETA1 ADRENERGIC RECEPTOR WITH STABILISING MUTATIONS AND BOUND ANTAGONIST CYANOPINDOLOL PDB File: | X-Ray Diffraction | 3.15 | 2011-06-08 | Moukhametzianov et al. Two Distinct Conformations of Helix 6 Observed in Antagonist-Bound Structures of a {Beta}1- Adrenergic Receptor. Proc.Natl.Acad.Sci.USA. 2011, 108, 8228. (PMID: 21540331) | |
 | Meleagris gallopavo (Wild turkey) (P07700) | TURKEY BETA1 ADRENERGIC RECEPTOR WITH STABILISING MUTATIONS AND BOUND ANTAGONIST IODOCYANOPINDOLOL PDB File: | X-Ray Diffraction | 3.65 | 2011-06-01 | Moukhametzianov et al. Two Distinct Conformations of Helix 6 Observed in Antagonist-Bound Structures of a {Beta}1- Adrenergic Receptor. Proc.Natl.Acad.Sci.USA. 2011, 108, 8228. (PMID: 21540331) | |
 | Meleagris gallopavo (Wild turkey) (P07700) | Thermostabilised turkey beta1 adrenergic receptor with 4-(piperazin-1- yl)-1H-indole bound (compound 19) PDB File: | X-Ray Diffraction | 2.8 | 2013-04-03 | Christopher et al. Biophysical Fragment Screening of the Beta1-Adrenergic Receptor: Identification of High Affinity Aryl Piperazine Leads Using Structure-Based Drug Design. J.Med.Chem. 2013, 56, 3446. (PMID: 23517028) | |
 | Meleagris gallopavo (Wild turkey) (P07700) | Thermostabilised turkey beta1 adrenergic receptor with 4-methyl-2-(piperazin-1-yl) quinoline bound PDB File: | X-Ray Diffraction | 2.7 | 2013-04-03 | Christopher et al. Biophysical Fragment Screening of the Beta1-Adrenergic Receptor: Identification of High Affinity Aryl Piperazine Leads Using Structure-Based Drug Design. J.Med.Chem. 2013, 56, 3446. (PMID: 23517028) | |
 | Meleagris gallopavo (Wild turkey) (P07700) | Turkey beta1 adrenergic receptor with stabilising mutations and bound biased agonist bucindolol PDB File: | X-Ray Diffraction | 3.2 | 2012-05-23 | Warne et al. Crystal Structures of a Stabilized Beta1-Adrenoceptor Bound to the Biased Agonists Bucindolol and Carvedilol. Structure. 2012, 20, 841. (PMID: 22579251) | |
 | Meleagris gallopavo (Wild turkey) (P07700) | Turkey beta1 adrenergic receptor with stabilising mutations and bound biased agonist carvedilol PDB File: | X-Ray Diffraction | 2.3 | 2012-05-23 | Warne et al. Crystal Structures of a Stabilized Beta1-Adrenoceptor Bound to the Biased Agonists Bucindolol and Carvedilol. Structure. 2012, 20, 841. (PMID: 22579251) | |
 | Meleagris gallopavo (Wild turkey) (P07700) | Ultra-thermostable beta1-adrenoceptor with cyanopindolol bound PDB File: | X-Ray Diffraction | 2.1 | 2014-04-02 | Miller-Gallacher et al. The 21 A Resolution Structure of Cyanopindolol-Bound Beta1- Adrenoceptor Identifies an Intramembrane Na+ Ion that Stabilises the Ligand-Free Receptor. Plos One. 2014, 9, 92727. (PMID: 24663151) | |
 | Meleagris gallopavo (Wild turkey) (P07700) | Oligomeic Turkey Beta1-Adrenergic G Protein-Coupled Receptor PDB File: | X-Ray Diffraction | 3.5 | 2013-02-27 | Huang et al. Crystal structure of oligomeric beta-1-adrenergic G protein-coupled receptors in ligand-free basal state. Nat.Struct.Mol.Biol. 2013, 20, 419. (PMID: 23435379) | |
 | Meleagris gallopavo (Wild turkey) (P07700) | thermostabilised beta1-adrenoceptor with rationally designed inverse agonist 7-methylcyanopindolol bound PDB File: | X-Ray Diffraction | 2.4 | 2015-09-30 | Sato et al. Pharmacological Analysis and Structure Determination of 7-Methylcyanopindolol-Bound Beta1-Adrenergic Receptor. Mol.Pharmacol. 2015, 88, 1024. (PMID: 26385885) | |
 | Meleagris gallopavo (Wild turkey) (P07700) | Ligand occupancy in crystal structure of beta1-adrenergic receptor previously submitted by Huang et al PDB File: | X-Ray Diffraction | 3.35 | 2015-12-23 | Leslie et al. Ligand occupancy in crystal structure of Beta1 adrenergic G protein coupled receptor. Nat.Struct.Mol.Biol. 2015, 22, 941. (PMID: 26643842) | |
 | Meleagris gallopavo (Wild turkey) (P07700) | ACTIVATED TURKEY BETA1 ADRENOCEPTOR WITH BOUND AGONIST ISOPRENALINE AND NANOBODY Nb80 PDB File: | X-Ray Diffraction | 2.8 | 2018-10-17 | Warne et al. Molecular basis for high-affinity agonist binding in GPCRs. Science. 2019. (PMID: 31072904) | |
 | Meleagris gallopavo (Wild turkey) (P07700) | ACTIVATED TURKEY BETA1 ADRENOCEPTOR WITH BOUND PARTIAL AGONIST DOBUTAMINE AND NANOBODY Nb6B9 PDB File: | X-Ray Diffraction | 2.7 | 2018-10-17 | Warne et al. Molecular basis for high-affinity agonist binding in GPCRs. Science. 2019. (PMID: 31072904) | |
 | Meleagris gallopavo (Wild turkey) (P07700) | ACTIVATED TURKEY BETA1 ADRENOCEPTOR WITH BOUND PARTIAL AGONIST SALBUTAMOL AND NANOBODY Nb6B9 PDB File: | X-Ray Diffraction | 2.76 | 2018-10-17 | Warne et al. Molecular basis for high-affinity agonist binding in GPCRs. Science. 2019. (PMID: 31072904) | |
 | Meleagris gallopavo (Wild turkey) (P07700) | ACTIVATED TURKEY BETA1 ADRENOCEPTOR WITH BOUND PARTIAL AGONIST XAMOTEROL AND NANOBODY Nb6B9 PDB File: | X-Ray Diffraction | 2.5 | 2018-10-17 | Warne et al. Molecular basis for high affinity agonist binding in GPCRs. Biorxiv. 2018. (PMID: N/A) | |
 | Meleagris gallopavo (Wild turkey) (P07700) | ACTIVATED TURKEY BETA1 ADRENOCEPTOR WITH BOUND WEAK PARTIAL AGONIST CYANOPINDOLOL AND NANOBODY Nb6B9 PDB File: | X-Ray Diffraction | 2.8 | 2018-10-17 | Warne et al. Molecular basis for high-affinity agonist binding in GPCRs. Science. 2019. (PMID: 31072904) | |
 | Meleagris gallopavo (Wild turkey) (P07700) | ACTIVATED TURKEY BETA1 ADRENOCEPTOR WITH BOUND AGONIST FORMOTEROL AND NANOBODY Nb80 PDB File: | X-Ray Diffraction | 2.7 | 2019-01-09 | Warne et al. Molecular basis for high affinity agonist binding in GPCRs. Biorxiv. 2018. (PMID: N/A) |
| PDB ID | Species (UniProt ID) | Description | Method | Resolution (Å) | Released | Reference | |
|---|---|---|---|---|---|---|---|
 | Homo sapiens (Human) (P07550) | Crystal structure of the human beta2 adrenoceptor PDB File: | X-Ray Diffraction | 3.4 | 2007-11-06 | Rasmussen et al. Crystal structure of the human beta2 adrenergic G-protein-coupled receptor. Nature. 2007, 450, 383. (PMID: 17952055) | |
 | Homo sapiens (Human) (P07550) | Crystal structure of the human beta2 adrenoceptor PDB File: | X-Ray Diffraction | 3.4 | 2007-11-06 | Rasmussen et al. Crystal structure of the human beta2 adrenergic G-protein-coupled receptor. Nature. 2007, 450, 383. (PMID: 17952055) | |
 | Homo sapiens (Human) (P07550) | High resolution crystal structure of human B2-adrenergic G protein-coupled receptor. PDB File: | X-Ray Diffraction | 2.4 | 2007-10-30 | Cherezov et al. High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Science. 2007, 318, 1258. (PMID: 17962520) | |
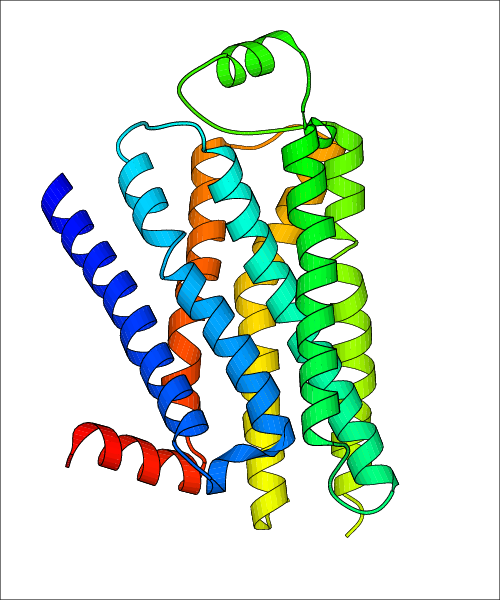 | Homo sapiens (Human) (P07550) | Cholesterol bound form of human beta2 adrenergic receptor. PDB File: | X-Ray Diffraction | 2.8 | 2008-06-17 | Hanson et al. A specific cholesterol binding site is established by the 28 A structure of the human beta2-adrenergic receptor. Structure. 2008, 16, 897. (PMID: 18547522) | |
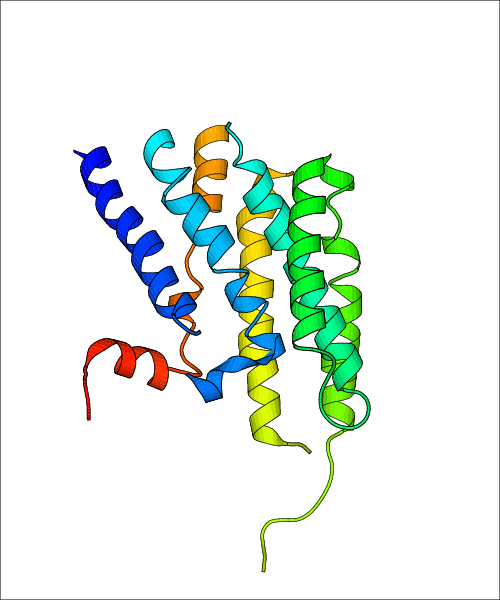 | Homo sapiens (Human) (P07550) | Crystal structure of a Methylated beta2 Adrenergic Receptor-Fab complex PDB File: | X-Ray Diffraction | 3.4 | 2010-02-16 | Bokoch et al. Ligand-specific regulation of the extracellular surface of a G-protein-coupled receptor. Nature. 2010, 463, 108. (PMID: 20054398) | |
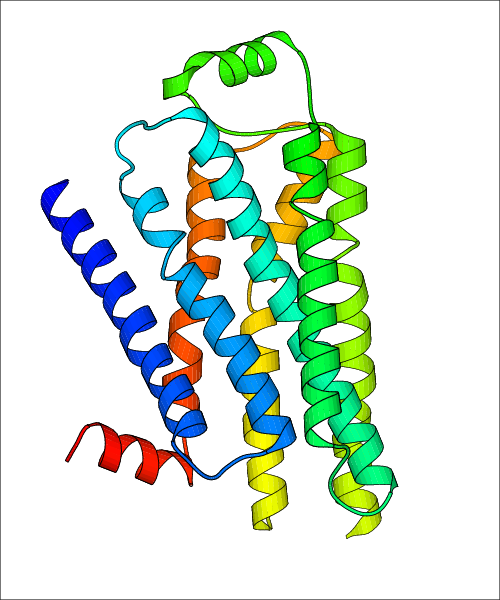 | Homo sapiens (Human) (P07550) | Crystal structure of the human beta2 adrenergic receptor in complex with the inverse agonist ICI 118,551 PDB File: | X-Ray Diffraction | 2.84 | 2010-08-11 | Wacker et al. Conserved Binding Mode of Human beta(2) Adrenergic Receptor Inverse Agonists and Antagonist Revealed by X-ray Crystallography. J.Am.Chem.Soc. 2010, 132, 11443. (PMID: 20669948) | |
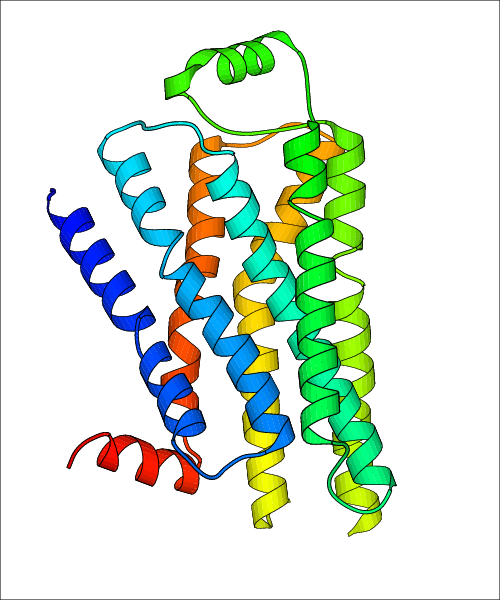 | Homo sapiens (Human) (P07550) | Crystal structure of the human beta2 adrenergic receptor in complex with a novel inverse agonist PDB File: | X-Ray Diffraction | 2.84 | 2010-08-11 | Wacker et al. Conserved Binding Mode of Human beta(2) Adrenergic Receptor Inverse Agonists and Antagonist Revealed by X-ray Crystallography. J.Am.Chem.Soc. 2010, 132, 11443. (PMID: 20669948) | |
 | Homo sapiens (Human) (P07550) | Crystal structure of the human beta2 adrenergic receptor in complex with the neutral antagonist alprenolol PDB File: | X-Ray Diffraction | 3.16 | 2010-08-11 | Wacker et al. Conserved Binding Mode of Human beta(2) Adrenergic Receptor Inverse Agonists and Antagonist Revealed by X-ray Crystallography. J.Am.Chem.Soc. 2010, 132, 11443. (PMID: 20669948) | |
 | Homo sapiens (Human) (P07550) | Structure of a nanobody-stabilized active state of the beta2 adrenoceptor PDB File: | X-Ray Diffraction | 3.5 | 2011-01-19 | Rasmussen et al. Structure of a nanobody-stabilized active state of the b2 adrenoceptor. Nature. 2011, 469, 175. (PMID: 21228869) | |
 | Homo sapiens (Human) (P07550) | Irreversible Agonist-Beta2 Adrenoceptor Complex PDB File: | X-Ray Diffraction | 3.5 | 2011-01-12 | Rosenbaum et al. Structure and function of an irreversible agonist-beta(2) adrenoceptor complex. Nature. 2011, 469, 236. (PMID: 21228876) | |
 | Homo sapiens (Human) (P07550) | Crystal structure of the beta2 adrenergic receptor-Gs protein complex PDB File: | X-Ray Diffraction | 3.2 | 2011-07-20 | Rasmussen et al. Crystal structure of the beta2 adrenergic receptor-Gs protein complex. Nature. 2011, 477, 549. (PMID: 21772288) | |
 | Homo sapiens (Human) (P07550) | N-Terminal T4 Lysozyme Fusion Facilitates Crystallization of a G Protein Coupled Receptor PDB File: | X-Ray Diffraction | 3.99 | 2012-10-24 | Zou et al. N-terminal t4 lysozyme fusion facilitates crystallization of a g protein coupled receptor. Plos One. 2012, 7, e46039. (PMID: 23056231) | |
 | Homo sapiens (Human) (P07550) | Structure of beta2 adrenoceptor bound to BI167107 and an engineered nanobody PDB File: | X-Ray Diffraction | 2.79 | 2013-09-25 | Ring et al. Adrenaline-activated structure of beta 2-adrenoceptor stabilized by an engineered nanobody. Nature. 2013, 502, 575. (PMID: 24056936) | |
 | Homo sapiens (Human) (P07550) | Structure of beta2 adrenoceptor bound to hydroxybenzylisoproterenol and an engineered nanobody PDB File: | X-Ray Diffraction | 3.1 | 2013-09-25 | Ring et al. Adrenaline-activated structure of beta 2-adrenoceptor stabilized by an engineered nanobody. Nature. 2013, 502, 575. (PMID: 24056936) | |
 | Homo sapiens (Human) (P07550) | Structure of beta2 adrenoceptor bound to adrenaline and an engineered nanobody PDB File: | X-Ray Diffraction | 3.2 | 2013-09-25 | Ring et al. Adrenaline-activated structure of beta 2-adrenoceptor stabilized by an engineered nanobody. Nature. 2013, 502, 575. (PMID: 24056936) | |
 | Homo sapiens (Human) (P07550) | Structure of beta2 adrenoceptor bound to a covalent agonist and an engineered nanobody PDB File: | X-Ray Diffraction | 3.3 | 2014-07-23 | Weichert et al. Covalent agonists for studying G protein-coupled receptor activation. Proc.Natl.Acad.Sci.USA. 2014, 111, 10744. (PMID: 25006259) | |
 | Homo sapiens (Human) (P07550) | In meso in situ serial X-ray crystallography structure of the Beta2-adrenergic receptor at 100 K PDB File: | X-Ray Diffraction | 2.48 | 2016-01-13 | Huang et al. In meso in situ serial X-ray crystallography of soluble and membrane proteins at cryogenic temperatures. Acta Crystallogr D Struct Biol. 2016, 72, 93. (PMID: 26894538) | |
 | Homo sapiens (Human) (P07550) | In meso X-ray crystallography structure of the Beta2-adrenergic receptor at 100 K PDB File: | X-Ray Diffraction | 3.8 | 2016-01-13 | Huang et al. In meso in situ serial X-ray crystallography of soluble and membrane proteins at cryogenic temperatures. Acta Crystallogr D Struct Biol. 2016, 72, 93. (PMID: 26894538) | |
 | Homo sapiens (Human) (P07550) | beta2AR-T4L - CIM PDB File: | X-Ray Diffraction | 3.2 | 2016-08-17 | Ma et al. The cubicon method for concentrating membrane proteins in the cubic mesophase. Nat Protoc. 2017, 12, 1745. (PMID: 28771236) | |
 | Homo sapiens (Human) (P07550) | Structure of beta2 adrenoceptor bound to carazolol and inactive-state stabilizing nanobody, Nb60 PDB File: | X-Ray Diffraction | 3.2 | 2016-07-13 | Staus et al. Allosteric nanobodies reveal the dynamic range and diverse mechanisms of G-protein-coupled receptor activation. Nature. 2016, 535, 448. (PMID: 27409812) | |
 | Homo sapiens (Human) (P07550) | Structure of beta2 adrenoceptor bound to carazolol and an intracellular allosteric antagonist PDB File: | X-Ray Diffraction | 2.7 | 2017-08-16 | Liu et al. Mechanism of intracellular allosteric beta 2AR antagonist revealed by X-ray crystal structure. Nature. 2017, 548, 480. (PMID: 28813418) | |
 | Homo sapiens (Human) (P07550) | Structure of beta2 adrenergic receptor fused to a Gs peptide PDB File: | X-Ray Diffraction | 3.7 | 2019-06-05 | Liu et al. Structural Insights into the Process of GPCR-G Protein Complex Formation. Cell. 2019, 177, 1243. (PMID: 31080070) | |
 | Homo sapiens (Human) (P07550) | Crystal structure of human beta2 adrenergic receptor bound to salmeterol and Nb71 PDB File: | X-Ray Diffraction | 2.96 | 2018-11-14 | Masureel et al. Structural insights into binding specificity, efficacy and bias of a beta2AR partial agonist. Nat. Chem. Biol. 2018, 14, 1059. (PMID: 30327561) | |
 | Homo sapiens (Human) (P07550) | Structure of beta2 adrenergic receptor bound to BI167107, Nanobody 6B9, and a positive allosteric modulator PDB File: | X-Ray Diffraction | 3.2 | 2019-06-26 | Liu et al. Mechanism of beta2AR regulation by an intracellular positive allosteric modulator. Science. 2019, 364, 1283. (PMID: 31249059) | |
 | Homo sapiens (Human) (P07550) | B2V2R-Gs protein subcomplex of a GPCR-G protein-beta-arrestin mega-complex PDB File: | Electron Microscopy | 3.8 | 2019-11-20 | Nguyen et al. Structure of an endosomal signaling GPCR-G protein-beta-arrestin megacomplex. Nat.Struct.Mol.Biol. 2019. (PMID: 31740855) | |
 | Homo sapiens (Human) (P07550) | XFEL beta2 AR structure by ligand exchange from Alprenolol to Alprenolol. PDB File: | X-Ray Diffraction | 2.8 | 2019-11-13 | Ishchenko et al. Toward G protein-coupled receptor structure-based drug design using X-ray lasers. Iucrj. 2019, 6, 1106. (PMID: 31709066) | |
 | Homo sapiens (Human) (P07550) | XFEL beta2 AR structure by ligand exchange from Alprenolol to Carazolol. PDB File: | X-Ray Diffraction | 3.4 | 2019-11-13 | Ishchenko et al. Toward G protein-coupled receptor structure-based drug design using X-ray lasers. Iucrj. 2019, 6, 1106. (PMID: 31709066) | |
 | Homo sapiens (Human) (P07550) | XFEL beta2 AR structure by ligand exchange from Alprenolol to Timolol. PDB File: | X-Ray Diffraction | 3.2 | 2019-11-13 | Ishchenko et al. Toward G protein-coupled receptor structure-based drug design using X-ray lasers. Iucrj. 2019, 6, 1106. (PMID: 31709066) | |
 | Homo sapiens (Human) (P07550) | XFEL beta2 AR structure by ligand exchange from Timolol to Alprenolol. PDB File: | X-Ray Diffraction | 2.4 | 2019-11-13 | Ishchenko et al. Toward G protein-coupled receptor structure-based drug design using X-ray lasers. Iucrj. 2019, 6, 1106. (PMID: 31709066) | |
 | Homo sapiens (Human) (P07550) | XFEL beta2 AR structure by ligand exchange from Timolol to Carvedilol. PDB File: | X-Ray Diffraction | 2.5 | 2019-11-13 | Ishchenko et al. Toward G protein-coupled receptor structure-based drug design using X-ray lasers. Iucrj. 2019, 6, 1106. (PMID: 31709066) | |
 | Homo sapiens (Human) (P07550) | XFEL beta2 AR structure by ligand exchange from Timolol to ICI-118551. PDB File: | X-Ray Diffraction | 2.6 | 2019-11-13 | Ishchenko et al. Toward G protein-coupled receptor structure-based drug design using X-ray lasers. Iucrj. 2019, 6, 1106. (PMID: 31709066) | |
 | Homo sapiens (Human) (P07550) | XFEL beta2 AR structure by ligand exchange from Timolol to Propranolol. PDB File: | X-Ray Diffraction | 2.9 | 2019-11-13 | Ishchenko et al. Toward G protein-coupled receptor structure-based drug design using X-ray lasers. Iucrj. 2019, 6, 1106. (PMID: 31709066) | |
 | Homo sapiens (Human) (P07550) | XFEL beta2 AR structure by ligand exchange from Timolol to Timolol. PDB File: | X-Ray Diffraction | 2.7 | 2019-11-13 | Ishchenko et al. Toward G protein-coupled receptor structure-based drug design using X-ray lasers. Iucrj. 2019, 6, 1106. (PMID: 31709066) |
| PDB ID | Species (UniProt ID) | Description | Method | Resolution (Å) | Released | Reference | |
|---|---|---|---|---|---|---|---|
 | Homo sapiens (Human) (P41597) | Structure of CC Chemokine Receptor 2 with Orthosteric and Allosteric Antagonists PDB File: | X-Ray Diffraction | 2.81 | 2016-12-14 | Zheng et al. Structure of CC chemokine receptor 2 with orthosteric and allosteric antagonists. Nature. 2016, 540, 458. (PMID: 27926736) | |
 | Homo sapiens (Human) (P41597) | CRYSTAL STRUCTURE OF CCR2A IN COMPLEX WITH MK-0812 PDB File: | X-Ray Diffraction | 3.3 | 2019-01-02 | Apel et al. Crystal Structure of CC Chemokine Receptor 2A in Complex with an Orthosteric Antagonist Provides Insights for the Design of Selective Antagonists. Structure. 2018. (PMID: 30581043) | |
 | Homo sapiens (Human) (P41597) | CRYSTAL STRUCTURE OF CCR2A IN COMPLEX WITH MK-0812 PDB File: | X-Ray Diffraction | 2.7 | 2019-01-02 | Apel et al. Crystal Structure of CC Chemokine Receptor 2A in Complex with an Orthosteric Antagonist Provides Insights for the Design of Selective Antagonists. Structure. 2018. (PMID: 30581043) |
| PDB ID | Species (UniProt ID) | Description | Method | Resolution (Å) | Released | Reference | |
|---|---|---|---|---|---|---|---|
 | Homo sapiens (Human) (P51681) | Crystal Structure of the CCR5 Chemokine Receptor PDB File: | X-Ray Diffraction | 2.71 | 2013-09-11 | Tan et al. Structure of the CCR5 chemokine receptor-HIV entry inhibitor maraviroc complex. Science. 2013, 341, 1387. (PMID: 24030490) | |
 | Homo sapiens (Human) (P51681) | Crystal Structure of CC Chemokine Receptor 5 (CCR5) in complex with high potency HIV entry inhibitor 5P7-CCL5 PDB File: | X-Ray Diffraction | 2.2 | 2017-06-28 | Zheng et al. Structure of CC Chemokine Receptor 5 with a Potent Chemokine Antagonist Reveals Mechanisms of Chemokine Recognition and Molecular Mimicry by HIV. Immunity. 2017, 46, 1005. (PMID: 28636951) | |
 | Homo sapiens (Human) (P51681) | The Crystal structure of Human Chemokine Receptor CCR5 in complex with compound 21 PDB File: | X-Ray Diffraction | 2.8 | 2018-10-24 | Peng et al. Structure-Based Design of 1-Heteroaryl-1,3-propanediamine Derivatives as a Novel Series of CC-Chemokine Receptor 5 Antagonists. J. Med. Chem. 2018, 61, 9621. (PMID: 30234300) | |
 | Homo sapiens (Human) (P51681) | The Crystal structure of Human Chemokine Receptor CCR5 in complex with compound 34 PDB File: | X-Ray Diffraction | 2.8 | 2018-10-24 | Peng et al. Structure-Based Design of 1-Heteroaryl-1,3-propanediamine Derivatives as a Novel Series of CC-Chemokine Receptor 5 Antagonists. J. Med. Chem. 2018, 61, 9621. (PMID: 30234300) | |
 | Homo sapiens (Human) (P51681) | Structural basis of coreceptor recognition by HIV-1 envelope spike PDB File: | Electron Microscopy | 3.9 | 2018-12-12 | Shaik et al. Structural basis of coreceptor recognition by HIV-1 envelope spike. Nature. 2018. (PMID: 30542158) | |
 | Homo sapiens (Human) (P51681) | Structural basis of coreceptor recognition by HIV-1 envelope spike PDB File: | Electron Microscopy | 4.5 | 2018-12-12 | Shaik et al. Structural basis of coreceptor recognition by HIV-1 envelope spike. Nature. 2018. (PMID: 30542158) |
| PDB ID | Species (UniProt ID) | Description | Method | Resolution (Å) | Released | Reference | |
|---|---|---|---|---|---|---|---|
 | Homo sapiens (Human) (P32248) | Structure of the human CC Chemokine Receptor 7 in complex with the intracellular allosteric antagonist Cmp2105 and the insertion protein Sialidase NanA PDB File: | X-Ray Diffraction | 2.1 | 2019-09-04 | Jaeger et al. Structural Basis for Allosteric Ligand Recognition in the Human CC Chemokine Receptor 7. Cell. 2019, 178, 1222. (PMID: 31442409) |
| PDB ID | Species (UniProt ID) | Description | Method | Resolution (Å) | Released | Reference | |
|---|---|---|---|---|---|---|---|
 | Homo sapiens (Human) (P51686) | Crystal structure of the human CC chemokine receptor type 9 (CCR9) in complex with vercirnon PDB File: | X-Ray Diffraction | 2.8 | 2016-12-07 | Oswald et al. Intracellular allosteric antagonism of the CCR9 receptor. Nature. 2016, 540, 462. (PMID: 27926729) |
| PDB ID | Species (UniProt ID) | Description | Method | Resolution (Å) | Released | Reference | |
|---|---|---|---|---|---|---|---|
 | Homo sapiens (Human) (P25024) | Structure of human CXCR1 in phospholipid bilayers PDB File: | Solid-State NMR | N/A | 2012-10-17 | Park et al. Structure of the chemokine receptor CXCR1 in phospholipid bilayers. Nature. 2012, 491, 779. (PMID: 23086146) |
| PDB ID | Species (UniProt ID) | Description | Method | Resolution (Å) | Released | Reference | |
|---|---|---|---|---|---|---|---|
 | Homo sapiens (Human) (P61073) | The 2.5 A structure of the CXCR4 chemokine receptor in complex with small molecule antagonist IT1t PDB File: | X-Ray Diffraction | 2.5 | 2010-10-27 | Wu et al. Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists. Science. 2010, 330, 1066. (PMID: 20929726) | |
 | Homo sapiens (Human) (P61073) | Crystal structure of the CXCR4 chemokine receptor in complex with a cyclic peptide antagonist CVX15 PDB File: | X-Ray Diffraction | 2.9 | 2010-10-27 | Wu et al. Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists. Science. 2010, 330, 1066. (PMID: 20929726) | |
 | Homo sapiens (Human) (P61073) | Crystal structure of the CXCR4 chemokine receptor in complex with a small molecule antagonist IT1t in I222 spacegroup PDB File: | X-Ray Diffraction | 3.2 | 2010-10-27 | Wu et al. Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists. Science. 2010, 330, 1066. (PMID: 20929726) | |
 | Homo sapiens (Human) (P61073) | Crystal structure of the CXCR4 chemokine receptor in complex with a small molecule antagonist IT1t in P1 spacegroup PDB File: | X-Ray Diffraction | 3.1 | 2010-10-27 | Wu et al. Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists. Science. 2010, 330, 1066. (PMID: 20929726) | |
 | Homo sapiens (Human) (P61073) | Crystal structure of the chemokine CXCR4 receptor in complex with a small molecule antagonist IT1t in P1 spacegroup PDB File: | X-Ray Diffraction | 3.1 | 2010-10-27 | Wu et al. Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists. Science. 2010, 330, 1066. (PMID: 20929726) | |
 | Homo sapiens (Human) (P61073) | Crystal structure of CXCR4 and viral chemokine antagonist vMIP-II complex (PSI Community Target) PDB File: | X-Ray Diffraction | 3.1 | 2015-02-11 | Qin et al. Structural biology Crystal structure of the chemokine receptor CXCR4 in complex with a viral chemokine. Science. 2015, 347, 1117. (PMID: 25612609) |
| PDB ID | Species (UniProt ID) | Description | Method | Resolution (Å) | Released | Reference | |
|---|---|---|---|---|---|---|---|
 | Homo sapiens (Human) (P21730) | Crystal structure of thermostabilised human C5a anaphylatoxin chemotactic receptor 1 (C5aR) in complex with NDT9513727 PDB File: | X-Ray Diffraction | 2.7 | 2018-01-10 | Robertson et al. Structure of the complement C5a receptor bound to the extra-helical antagonist NDT9513727. Nature. 2018, 553, 111. (PMID: 29300009) | |
 | Homo sapiens (Human) (P21730) | Crystal structure of human C5a receptor in complex with an orthosteric antagonist PMX53 and an allosteric antagonist NDT9513727 PDB File: | X-Ray Diffraction | 2.9 | 2018-05-30 | Liu et al. Orthosteric and allosteric action of the C5a receptor antagonists. Nat. Struct. Mol. Biol. 2018, 25, 472. (PMID: 29867214) | |
 | Homo sapiens (Human) (P21730) | Crystal structure of human C5a receptor in complex with an orthosteric antagonist PMX53 and an allosteric antagonist avacopan PDB File: | X-Ray Diffraction | 2.2 | 2018-05-30 | Liu et al. Orthosteric and allosteric action of the C5a receptor antagonists. Nat. Struct. Mol. Biol. 2018, 25, 472. (PMID: 29867214) |
| PDB ID | Species (UniProt ID) | Description | Method | Resolution (Å) | Released | Reference | |
|---|---|---|---|---|---|---|---|
 | Homo sapiens (Human) (P21554) | Crystal Structure of the Human Cannabinoid Receptor CB1 PDB File: | X-Ray Diffraction | 2.8 | 2016-11-02 | Hua et al. Crystal Structure of the Human Cannabinoid Receptor CB1. Cell. 2016, 167, 750. (PMID: 27768894) | |
 | Homo sapiens (Human) (P21554) | High-resolution crystal structure of the human CB1 cannabinoid receptor PDB File: | X-Ray Diffraction | 2.6 | 2016-12-07 | Shao et al. High-resolution crystal structure of the human CB1 cannabinoid receptor. Nature. 2016. (PMID: 27851727) | |
 | Homo sapiens (Human) (P21554) | Crystal structure of the human CB1 in complex with agonist AM841 PDB File: | X-Ray Diffraction | 2.95 | 2017-07-12 | Hua et al. Crystal structures of agonist-bound human cannabinoid receptor CB1. Nature. 2017, 547, 468. (PMID: 28678776) | |
 | Homo sapiens (Human) (P21554) | Crystal structure of the human CB1 in complex with agonist AM11542 PDB File: | X-Ray Diffraction | 2.8 | 2017-07-12 | Hua et al. Crystal structures of agonist-bound human cannabinoid receptor CB1. Nature. 2017, 547, 468. (PMID: 28678776) | |
 | Homo sapiens (Human) (P21554) | Crystal Structure of protein1 PDB File: | X-Ray Diffraction | 3.24 | 2019-10-23 | Shao et al. Structure of an allosteric modulator bound to the CB1 cannabinoid receptor. Nat.Chem.Biol. 2019. (PMID: 31659318) | |
 | Homo sapiens (Human) (P21554) | Cannabinoid Receptor 1-G Protein Complex PDB File: | Electron Microscopy | 3.0 | 2019-01-30 | Krishna Kumar et al. Structure of a Signaling Cannabinoid Receptor 1-G Protein Complex. Cell. 2018. (PMID: 30639101) |
| PDB ID | Species (UniProt ID) | Description | Method | Resolution (Å) | Released | Reference | |
|---|---|---|---|---|---|---|---|
 | Homo sapiens (Human) (P34972) | Crystal structure of human G protein coupled receptor PDB File: | X-Ray Diffraction | 2.8 | 2019-01-30 | Li et al. Crystal Structure of the Human Cannabinoid Receptor CB2. Cell. 2019. (PMID: 30639103) |
| PDB ID | Species (UniProt ID) | Description | Method | Resolution (Å) | Released | Reference | |
|---|---|---|---|---|---|---|---|
 | Homo sapiens (Human) (Q9Y271) | Crystal structure of cysteinyl leukotriene receptor 1 in complex with pranlukast PDB File: | X-Ray Diffraction | 2.7 | 2019-10-30 | Luginina et al. Structure-based mechanism of cysteinyl leukotriene receptor inhibition by antiasthmatic drugs. Sci Adv. 2019, 5, eaax2518. (PMID: 31633023) | |
 | Homo sapiens (Human) (Q9Y271) | XFEL crystal structure of the human cysteinyl leukotriene receptor 1 in complex with zafirlukast PDB File: | X-Ray Diffraction | 2.53 | 2019-10-30 | Luginina et al. Structure-based mechanism of cysteinyl leukotriene receptor inhibition by antiasthmatic drugs. Sci Adv. 2019, 5, eaax2518. (PMID: 31633023) |
| PDB ID | Species (UniProt ID) | Description | Method | Resolution (Å) | Released | Reference | |
|---|---|---|---|---|---|---|---|
 | Homo sapiens (Human) (Q9NS75) | Crystal structure of the human cysteinyl leukotriene receptor 2 in complex with ONO-2570366 (C2221 space group) PDB File: | X-Ray Diffraction | 2.43 | 2019-12-11 | Gusach et al. Structural basis of ligand selectivity and disease mutations in cysteinyl leukotriene receptors. Nat Commun. 2019, 10, 5573. (PMID: 31811124) | |
 | Homo sapiens (Human) (Q9NS75) | Crystal structure of the human cysteinyl leukotriene receptor 2 in complex with ONO-2570366 (F222 space group) PDB File: | X-Ray Diffraction | 2.43 | 2019-12-11 | Gusach et al. Structural basis of ligand selectivity and disease mutations in cysteinyl leukotriene receptors. Nat Commun. 2019, 10, 5573. (PMID: 31811124) | |
 | Homo sapiens (Human) (Q9NS75) | Crystal structure of the human cysteinyl leukotriene receptor 2 in complex with ONO-2080365 PDB File: | X-Ray Diffraction | 2.7 | 2019-12-11 | Gusach et al. Structural basis of ligand selectivity and disease mutations in cysteinyl leukotriene receptors. Nat Commun. 2019, 10, 5573. (PMID: 31811124) | |
 | Homo sapiens (Human) (Q9NS75) | Crystal structure of the human cysteinyl leukotriene receptor 2 in complex with ONO-2770372 PDB File: | X-Ray Diffraction | 2.73 | 2019-12-11 | Gusach et al. Structural basis of ligand selectivity and disease mutations in cysteinyl leukotriene receptors. Nat Commun. 2019, 10, 5573. (PMID: 31811124) |
| PDB ID | Species (UniProt ID) | Description | Method | Resolution (Å) | Released | Reference | |
|---|---|---|---|---|---|---|---|
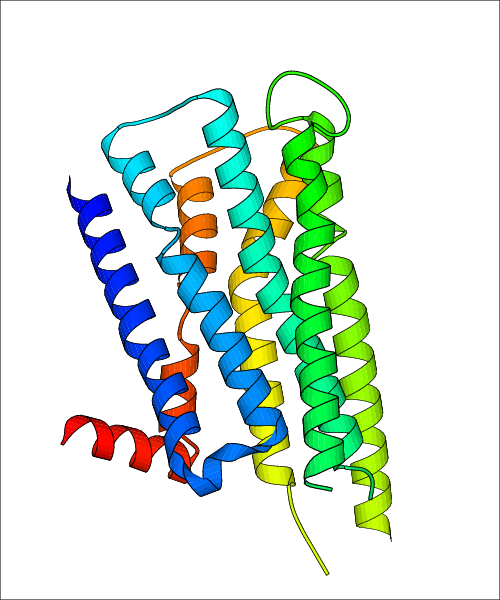 | Homo sapiens (Human) (P14416) | Structure of the D2 Dopamine Receptor Bound to the Atypical Antipsychotic Drug Risperidone PDB File: | X-Ray Diffraction | 2.87 | 2018-03-14 | Wang et al. Structure of the D2 dopamine receptor bound to the atypical antipsychotic drug risperidone. Nature. 2018, 555, 269. (PMID: 29466326) |
| PDB ID | Species (UniProt ID) | Description | Method | Resolution (Å) | Released | Reference | |
|---|---|---|---|---|---|---|---|
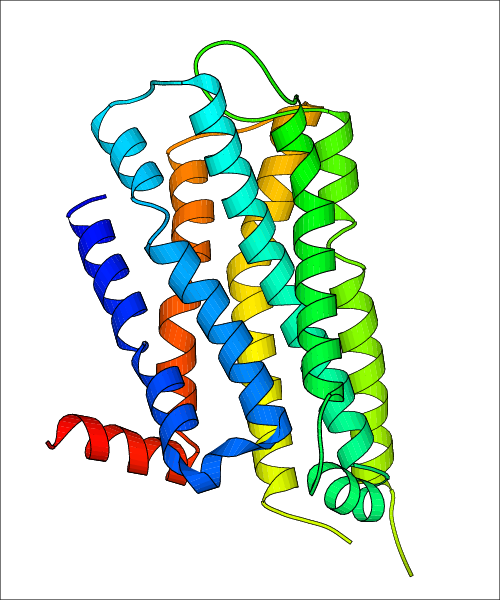 | Homo sapiens (Human) (P35462) | Structure of the human dopamine D3 receptor in complex with eticlopride PDB File: | X-Ray Diffraction | 2.89 | 2010-11-03 | Chien et al. Structure of the human dopamine d3 receptor in complex with a d2/d3 selective antagonist. Science. 2010, 330, 1091. (PMID: 21097933) |
| PDB ID | Species (UniProt ID) | Description | Method | Resolution (Å) | Released | Reference | |
|---|---|---|---|---|---|---|---|
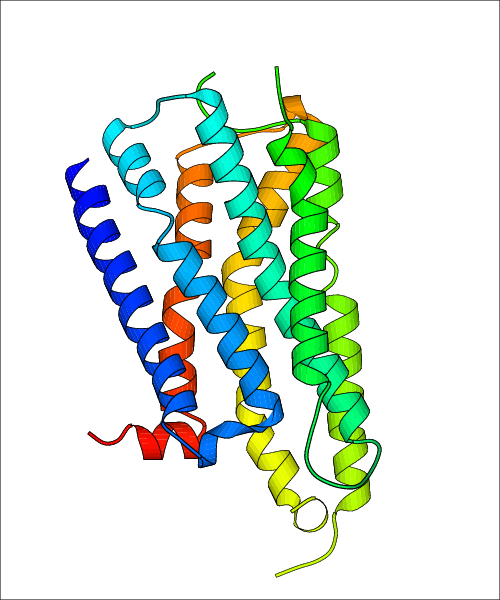 | Homo sapiens (Human) (P21917) | Structure of the human D4 Dopamine receptor in complex with Nemonapride PDB File: | X-Ray Diffraction | 1.96 | 2017-10-18 | Wang et al. D4 dopamine receptor high-resolution structures enable the discovery of selective agonists. Science. 2017, 358, 381. (PMID: 29051383) | |
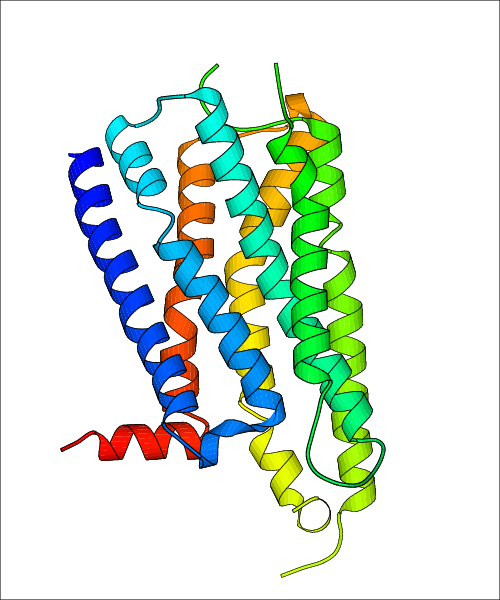 | Homo sapiens (Human) (P21917) | Structure of the sodium-bound human D4 Dopamine receptor in complex with Nemonapride PDB File: | X-Ray Diffraction | 2.14 | 2017-10-18 | Wang et al. D4 dopamine receptor high-resolution structures enable the discovery of selective agonists. Science. 2017, 358, 381. (PMID: 29051383) | |
 | Mus musculus (Mouse) (P51436) | Crystal structure of dopamine receptor D4 bound to the subtype-selective ligand, L745870 PDB File: | X-Ray Diffraction | 3.5 | 2019-12-04 | Zhou et al. Crystal structure of dopamine receptor D4 bound to the subtype selective ligand, L745870. Elife. 2019, 8, . (PMID: 31750832) |
| PDB ID | Species (UniProt ID) | Description | Method | Resolution (Å) | Released | Reference | |
|---|---|---|---|---|---|---|---|
 | Mus musculus (Mouse) (P32300) | Structure of the delta opioid receptor bound to naltrindole PDB File: | X-Ray Diffraction | 3.4 | 2012-05-16 | Granier et al. Structure of the delta opioid receptor bound to naltrindole. Nature. 2012, 485, 400. (PMID: 22596164) | |
 | Homo sapiens (Human) (P41143) | 1.8 A Structure of the human delta opioid 7TM receptor (PSI Community Target) PDB File: | X-Ray Diffraction | 1.8 | 2013-12-25 | Fenalti et al. Molecular control of delta-opioid receptor signalling. Nature. 2014, 506, 191. (PMID: 24413399) | |
 | Homo sapiens (Human) (P41143) | Synchrotron structure of the human delta opioid receptor in complex with a bifunctional peptide (PSI community target) PDB File: | X-Ray Diffraction | 3.28 | 2015-01-14 | Fenalti et al. Structural basis for bifunctional peptide recognition at human delta-opioid receptor. Nat.Struct.Mol.Biol. 2015, 22, 265. (PMID: 25686086) | |
 | Homo sapiens (Human) (P41143) | XFEL structure of the human delta opioid receptor in complex with a bifunctional peptide PDB File: | X-Ray Diffraction | 2.7 | 2015-01-14 | Fenalti et al. Structural basis for bifunctional peptide recognition at human delta-opioid receptor. Nat.Struct.Mol.Biol. 2015, 22, 265. (PMID: 25686086) |
| PDB ID | Species (UniProt ID) | Description | Method | Resolution (Å) | Released | Reference | |
|---|---|---|---|---|---|---|---|
 | Homo sapiens (Human) (P24530) | Human endothelin receptor type-B in complex with ET-1 PDB File: | X-Ray Diffraction | 2.8 | 2016-09-07 | Shihoya et al. Activation mechanism of endothelin ETB receptor by endothelin-1. Nature. 2016, 537, 363. (PMID: 27595334) | |
 | Homo sapiens (Human) (P24530) | Human endothelin receptor type-B in the ligand-free form PDB File: | X-Ray Diffraction | 2.5 | 2016-09-07 | Shihoya et al. Activation mechanism of endothelin ETB receptor by endothelin-1. Nature. 2016, 537, 363. (PMID: 27595334) | |
 | Homo sapiens (Human) (P24530) | Human endothelin receptor type-B in complex with antagonist K-8794 PDB File: | X-Ray Diffraction | 2.2 | 2017-08-16 | Shihoya et al. X-ray structures of endothelin ETB receptor bound to clinical antagonist bosentan and its analog. Nat. Struct. Mol. Biol. 2017, 24, 758. (PMID: 28805809) | |
 | Homo sapiens (Human) (P24530) | Human endothelin receptor type-B in complex with antagonist bosentan PDB File: | X-Ray Diffraction | 3.6 | 2017-08-16 | Shihoya et al. X-ray structures of endothelin ETB receptor bound to clinical antagonist bosentan and its analog. Nat. Struct. Mol. Biol. 2017, 24, 758. (PMID: 28805809) | |
 | Homo sapiens (Human) (P24530) | Crystal Structure of human ETB receptor in complex with Endothelin-3 PDB File: | X-Ray Diffraction | 2.0 | 2018-11-21 | Shihoya et al. Crystal structures of human ETBreceptor provide mechanistic insight into receptor activation and partial activation. Nat Commun. 2018, 9, 4711. (PMID: 30413709) | |
 | Homo sapiens (Human) (P24530) | Crystal Structure of human ETB receptor in complex with IRL1620 PDB File: | X-Ray Diffraction | 2.7 | 2018-11-21 | Shihoya et al. Crystal structures of human ETBreceptor provide mechanistic insight into receptor activation and partial activation. Nat Commun. 2018, 9, 4711. (PMID: 30413709) | |
 | Homo sapiens (Human) (P24530) | Human endothelin receptor type-B in complex with inverse agonist IRL2500 PDB File: | X-Ray Diffraction | 2.7 | 2019-07-17 | Nagiri et al. Crystal structure of human endothelin ETBreceptor in complex with peptide inverse agonist IRL2500. Commun Biol. 2019, 2, 236. (PMID: 31263780) |
| PDB ID | Species (UniProt ID) | Description | Method | Resolution (Å) | Released | Reference | |
|---|---|---|---|---|---|---|---|
 | Homo sapiens (Human) (O14842) | Crystal structure of Human GPR40 bound to allosteric agonist TAK-875 PDB File: | X-Ray Diffraction | 2.33 | 2014-07-16 | Srivastava et al. High-resolution structure of the human GPR40 receptor bound to allosteric agonist TAK-875. Nature. 2014, 513, 124. (PMID: 25043059) | |
 | Homo sapiens (Human) (O14842) | The extra-helical binding site of GPR40 and the structural basis for allosteric agonism and incretin stimulation PDB File: | X-Ray Diffraction | 2.76 | 2018-05-02 | Ho et al. Structural basis for GPR40 allosteric agonism and incretin stimulation. Nat Commun. 2018, 9, 1645. (PMID: 29695780) | |
 | Homo sapiens (Human) (O14842) | GPR40 in complex with partial agonist MK-8666 PDB File: | X-Ray Diffraction | 2.2 | 2017-06-07 | Lu et al. Structural basis for the cooperative allosteric activation of the free fatty acid receptor GPR40. Nat. Struct. Mol. Biol. 2017, 24, 570. (PMID: 28581512) | |
 | Homo sapiens (Human) (O14842) | GPR40 in complex with AgoPAM AP8 and partial agonist MK-8666 PDB File: | X-Ray Diffraction | 3.22 | 2017-06-07 | Lu et al. Structural basis for the cooperative allosteric activation of the free fatty acid receptor GPR40. Nat. Struct. Mol. Biol. 2017, 24, 570. (PMID: 28581512) |
| PDB ID | Species (UniProt ID) | Description | Method | Resolution (Å) | Released | Reference | |
|---|---|---|---|---|---|---|---|
 | Human cytomegalovirus (strain AD169) (HHV-5) (P69332) | Structure of a nanobody-bound viral GPCR bound to human chemokine CX3CL1 PDB File: | X-Ray Diffraction | 2.89 | 2015-03-04 | Burg et al. Structural biology Structural basis for chemokine recognition and activation of a viral G protein-coupled receptor. Science. 2015, 347, 1113. (PMID: 25745166) | |
 | Human cytomegalovirus (strain AD169) (HHV-5) (P69332) | Structure of a viral GPCR bound to human chemokine CX3CL1 PDB File: | X-Ray Diffraction | 3.8 | 2015-03-04 | Burg et al. Structural biology Structural basis for chemokine recognition and activation of a viral G protein-coupled receptor. Science. 2015, 347, 1113. (PMID: 25745166) | |
 | Human cytomegalovirus (strain Towne) (HHV-5) (P69333) | Ligand-free US28 with stabilizing intracellular nanobody PDB File: | X-Ray Diffraction | 3.51 | 2018-06-13 | Miles et al. Viral GPCR US28 can signal in response to chemokine agonists of nearly unlimited structural degeneracy. Elife. 2018, 7, . (PMID: 29882741) | |
 | Human cytomegalovirus (strain Towne) (HHV-5) (P69333) | US28 bound to engineered chemokine CX3CL1.35 and nanobodies PDB File: | X-Ray Diffraction | 3.5 | 2018-06-13 | Miles et al. Viral GPCR US28 can signal in response to chemokine agonists of nearly unlimited structural degeneracy. Elife. 2018, 7, . (PMID: 29882741) |
| PDB ID | Species (UniProt ID) | Description | Method | Resolution (Å) | Released | Reference | |
|---|---|---|---|---|---|---|---|
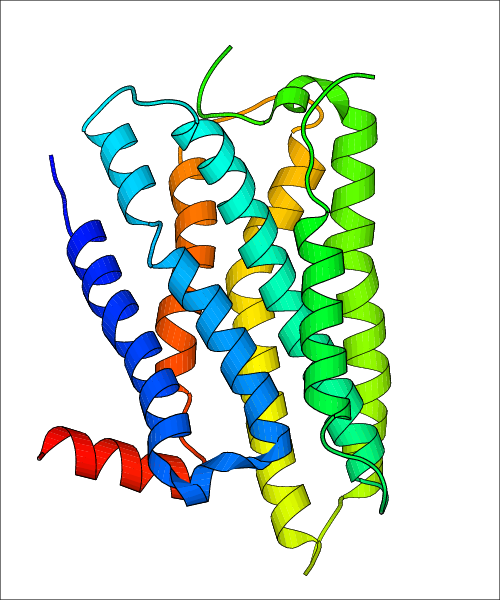 | Homo sapiens (Human) (P35367) | Structure of the human histamine H1 receptor in complex with doxepin PDB File: | X-Ray Diffraction | 3.1 | 2011-06-15 | Shimamura et al. Structure of the human histamine H1 receptor complex with doxepin. Nature. 2011, 475, 65. (PMID: 21697825) |
| PDB ID | Species (UniProt ID) | Description | Method | Resolution (Å) | Released | Reference | |
|---|---|---|---|---|---|---|---|
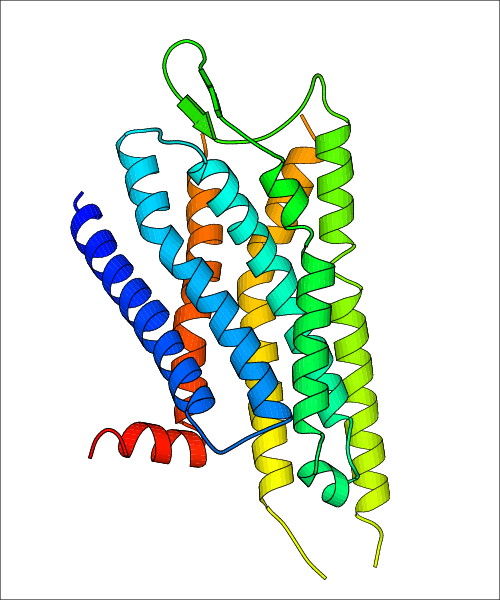 | Homo sapiens (Human) (P41145) | Structure of the human kappa opioid receptor in complex with JDTic PDB File: | X-Ray Diffraction | 2.9 | 2012-03-21 | Wu et al. Structure of the human kappa-opioid receptor in complex with JDTic. Nature. 2012, 485, 327. (PMID: 22437504) | |
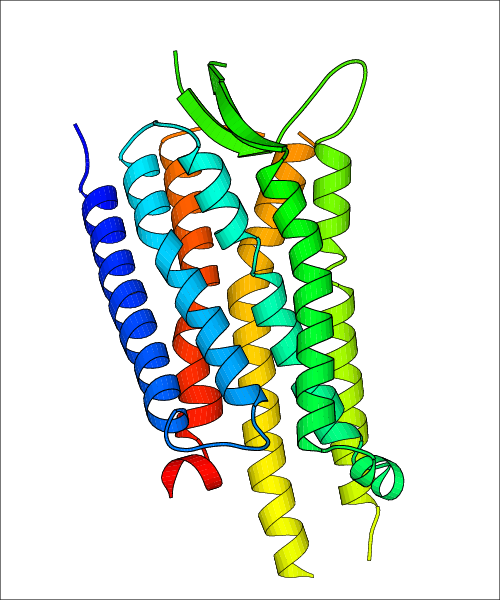 | Homo sapiens (Human) (P41145) | Crystal Structure of a nanobody-stabilized active state of the kappa-opioid receptor PDB File: | X-Ray Diffraction | 3.1 | 2018-01-17 | Che et al. Structure of the Nanobody-Stabilized Active State of the Kappa Opioid Receptor. Cell. 2018, 172, 55. (PMID: 29307491) |
| PDB ID | Species (UniProt ID) | Description | Method | Resolution (Å) | Released | Reference | |
|---|---|---|---|---|---|---|---|
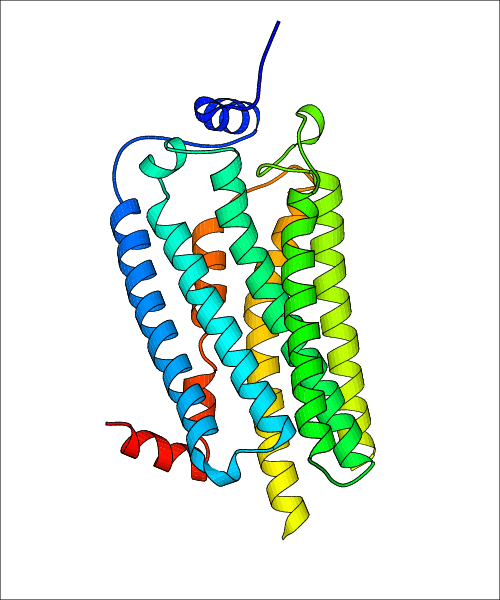 | Homo sapiens (Human) (Q92633) | Crystal Structure of Human Lysophosphatidic Acid Receptor 1 in complex with ONO9780307 PDB File: | X-Ray Diffraction | 3.0 | 2015-06-03 | Chrencik et al. Crystal Structure of Antagonist Bound Human Lysophosphatidic Acid Receptor 1. Cell. 2015, 161, 1633. (PMID: 26091040) | |
 | Homo sapiens (Human) (Q92633) | Crystal Structure of Human Lysophosphatidic Acid Receptor 1 in complex with ONO-9910539 PDB File: | X-Ray Diffraction | 2.9 | 2015-06-03 | Chrencik et al. Crystal Structure of Antagonist Bound Human Lysophosphatidic Acid Receptor 1. Cell. 2015, 161, 1633. (PMID: 26091040) | |
 | Homo sapiens (Human) (Q92633) | Crystal Structure of Human Lysophosphatidic Acid Receptor 1 in complex with ONO-3080573 PDB File: | X-Ray Diffraction | 2.9 | 2015-06-03 | Chrencik et al. Crystal Structure of Antagonist Bound Human Lysophosphatidic Acid Receptor 1. Cell. 2015, 161, 1633. (PMID: 26091040) |
| PDB ID | Species (UniProt ID) | Description | Method | Resolution (Å) | Released | Reference | |
|---|---|---|---|---|---|---|---|
 | Homo sapiens (Human) (P48039) | XFEL crystal structure of human melatonin receptor MT1 in complex with ramelteon PDB File: | X-Ray Diffraction | 2.8 | 2019-04-24 | Stauch et al. Structural basis of ligand recognition at the human MT1melatonin receptor. Nature. 2019. (PMID: 31019306) | |
 | Homo sapiens (Human) (P48039) | XFEL crystal structure of human melatonin receptor MT1 in complex with 2-phenylmelatonin PDB File: | X-Ray Diffraction | 2.9 | 2019-04-24 | Stauch et al. Structural basis of ligand recognition at the human MT1melatonin receptor. Nature. 2019. (PMID: 31019306) | |
 | Homo sapiens (Human) (P48039) | XFEL crystal structure of human melatonin receptor MT1 in complex with 2-iodomelatonin PDB File: | X-Ray Diffraction | 3.2 | 2019-04-24 | Stauch et al. Structural basis of ligand recognition at the human MT1melatonin receptor. Nature. 2019. (PMID: 31019306) | |
 | Homo sapiens (Human) (P48039) | XFEL crystal structure of human melatonin receptor MT1 in complex with agomelatine PDB File: | X-Ray Diffraction | 3.2 | 2019-04-24 | Stauch et al. Structural basis of ligand recognition at the human MT1melatonin receptor. Nature. 2019. (PMID: 31019306) | |
 | Homo sapiens (Human) (P48039) | XFEL MT1R structure by ligand exchange from agomelatine to 2-phenylmelatonin. PDB File: | X-Ray Diffraction | 3.3 | 2019-11-13 | Ishchenko et al. Toward G protein-coupled receptor structure-based drug design using X-ray lasers. Iucrj. 2019, 6, 1106. (PMID: 31709066) |
| PDB ID | Species (UniProt ID) | Description | Method | Resolution (Å) | Released | Reference | |
|---|---|---|---|---|---|---|---|
 | Homo sapiens (Human) (P49286) | XFEL crystal structure of human melatonin receptor MT2 in complex with 2-phenylmelatonin PDB File: | X-Ray Diffraction | 2.8 | 2019-04-24 | Johansson et al. XFEL structures of the human MT2melatonin receptor reveal the basis of subtype selectivity. Nature. 2019. (PMID: 31019305) | |
 | Homo sapiens (Human) (P49286) | XFEL crystal structure of human melatonin receptor MT2 (H208A) in complex with 2-phenylmelatonin PDB File: | X-Ray Diffraction | 3.2 | 2019-04-24 | Johansson et al. XFEL structures of the human MT2melatonin receptor reveal the basis of subtype selectivity. Nature. 2019. (PMID: 31019305) | |
 | Homo sapiens (Human) (P49286) | XFEL crystal structure of human melatonin receptor MT2 (N86D) in complex with 2-phenylmelatonin PDB File: | X-Ray Diffraction | 3.1 | 2019-04-24 | Johansson et al. XFEL structures of the human MT2melatonin receptor reveal the basis of subtype selectivity. Nature. 2019. (PMID: 31019305) | |
 | Homo sapiens (Human) (P49286) | XFEL crystal structure of human melatonin receptor MT2 in complex with ramelteon PDB File: | X-Ray Diffraction | 3.3 | 2019-04-24 | Johansson et al. XFEL structures of the human MT2melatonin receptor reveal the basis of subtype selectivity. Nature. 2019. (PMID: 31019305) |
| PDB ID | Species (UniProt ID) | Description | Method | Resolution (Å) | Released | Reference | |
|---|---|---|---|---|---|---|---|
 | Mus musculus (Mouse) (P42866) | Crystal structure of the mu-opioid receptor bound to a morphinan antagonist PDB File: | X-Ray Diffraction | 2.8 | 2012-03-21 | Manglik et al. Crystal structure of the {mu}-opioid receptor bound to a morphinan antagonist. Nature. 2012, 485, 321. (PMID: 22437502) | |
 | Mus musculus (Mouse) (P42866) | Crystal structure of active mu-opioid receptor bound to the agonist BU72 PDB File: | X-Ray Diffraction | 2.1 | 2015-08-05 | Huang et al. Structural insights into mu-opioid receptor activation. Nature. 2015, 524, 315. (PMID: 26245379) | |
 | Mus musculus (Mouse) (P42866) | Mu Opioid Receptor-Gi Protein Complex PDB File: | Electron Microscopy | 3.5 | 2018-06-13 | Koehl et al. Structure of the μ-opioid receptor-Giprotein complex. Nature. 2018. (PMID: 29899455) | |
 | Mus musculus (Mouse) (P42866) | Mu Opioid Receptor-Gi Protein Complex PDB File: | Electron Microscopy | 3.5 | 2018-06-13 | Koehl et al. Structure of the μ-opioid receptor-Giprotein complex. Nature. 2018. (PMID: 29899455) |
| PDB ID | Species (UniProt ID) | Description | Method | Resolution (Å) | Released | Reference | |
|---|---|---|---|---|---|---|---|
 | Homo sapiens (Human) (P11229) | Structure of the human M1 muscarinic acetylcholine receptor bound to antagonist Tiotropium PDB File: | X-Ray Diffraction | 2.7 | 2016-03-09 | Thal et al. Crystal structures of the M1 and M4 muscarinic acetylcholine receptors. Nature. 2016, 531, 335. (PMID: 26958838) | |
 | Homo sapiens (Human) (P11229) | Muscarinic acetylcholine receptor 1-G11 protein complex PDB File: | Electron Microscopy | 3.3 | 2019-05-08 | Maeda et al. Structures of the M1 and M2 muscarinic acetylcholine receptor/G-protein complexes. Science. 2019, 364, 552. (PMID: 31073061) |
| PDB ID | Species (UniProt ID) | Description | Method | Resolution (Å) | Released | Reference | |
|---|---|---|---|---|---|---|---|
 | Homo sapiens (Human) (P08172) | Structure of the human M2 muscarinic acetylcholine receptor bound to an antagonist PDB File: | X-Ray Diffraction | 3.0 | 2012-02-01 | Haga et al. Structure of the human M2 muscarinic acetylcholine receptor bound to an antagonist. Nature. 2012, 482, 547. (PMID: 22278061) | |
 | Homo sapiens (Human) (P08172) | Structure of active human M2 muscarinic acetylcholine receptor bound to the agonist iperoxo PDB File: | X-Ray Diffraction | 3.5 | 2013-11-27 | Kruse et al. Activation and allosteric modulation of a muscarinic acetylcholine receptor. Nature. 2013, 504, 101. (PMID: 24256733) | |
 | Homo sapiens (Human) (P08172) | Structure of active human M2 muscarinic acetylcholine receptor bound to the agonist iperoxo and allosteric modulator LY2119620 PDB File: | X-Ray Diffraction | 3.7 | 2013-11-27 | Kruse et al. Activation and allosteric modulation of a muscarinic acetylcholine receptor. Nature. 2013, 504, 101. (PMID: 24256733) | |
 | Homo sapiens (Human) (P08172) | Crystal structure of rationally thermostabilized M2 muscarinic acetylcholine receptor bound with NMS (Hg-derivative) PDB File: | X-Ray Diffraction | 2.5 | 2018-11-21 | Suno et al. Structural insights into the subtype-selective antagonist binding to the M2muscarinic receptor. Nat. Chem. Biol. 2018, 14, 1150. (PMID: 30420692) | |
 | Homo sapiens (Human) (P08172) | Crystal structure of rationally thermostabilized M2 muscarinic acetylcholine receptor bound with QNB PDB File: | X-Ray Diffraction | 2.6 | 2018-11-21 | Suno et al. Structural insights into the subtype-selective antagonist binding to the M2muscarinic receptor. Nat. Chem. Biol. 2018, 14, 1150. (PMID: 30420692) | |
 | Homo sapiens (Human) (P08172) | Crystal structure of M2 muscarinic acetylcholine receptor bound with NMS PDB File: | X-Ray Diffraction | 3.0 | 2018-11-21 | Suno et al. Structural insights into the subtype-selective antagonist binding to the M2muscarinic receptor. Nat. Chem. Biol. 2018, 14, 1150. (PMID: 30420692) | |
 | Homo sapiens (Human) (P08172) | Crystal structure of rationally thermostabilized M2 muscarinic acetylcholine receptor bound with AF-DX 384 PDB File: | X-Ray Diffraction | 2.95 | 2018-11-21 | Suno et al. Structural insights into the subtype-selective antagonist binding to the M2muscarinic receptor. Nat. Chem. Biol. 2018, 14, 1150. (PMID: 30420692) | |
 | Homo sapiens (Human) (P08172) | Crystal structure of rationally thermostabilized M2 muscarinic acetylcholine receptor bound with NMS PDB File: | X-Ray Diffraction | 2.3 | 2018-11-21 | Suno et al. Structural insights into the subtype-selective antagonist binding to the M2muscarinic receptor. Nat. Chem. Biol. 2018, 14, 1150. (PMID: 30420692) | |
 | Homo sapiens (Human) (P08172) | Muscarinic acetylcholine receptor 2-Go complex PDB File: | Electron Microscopy | 3.6 | 2019-05-08 | Maeda et al. Structures of the M1 and M2 muscarinic acetylcholine receptor/G-protein complexes. Science. 2019, 364, 552. (PMID: 31073061) |
| PDB ID | Species (UniProt ID) | Description | Method | Resolution (Å) | Released | Reference | |
|---|---|---|---|---|---|---|---|
 | Rattus norvegicus (Rat) (P08483) | Structure of the M3 Muscarinic Acetylcholine Receptor PDB File: | X-Ray Diffraction | 3.4 | 2012-02-22 | Kruse et al. Structure and dynamics of the M3 muscarinic acetylcholine receptor. Nature. 2012, 482, 552. (PMID: 22358844) | |
 | Rattus norvegicus (Rat) (P08483) | Structure of the M3 muscarinic acetylcholine receptor bound to the antagonist tiotropium crystallized with disulfide-stabilized T4 lysozyme (dsT4L) PDB File: | X-Ray Diffraction | 3.57 | 2014-11-26 | Thorsen et al. Modified T4 Lysozyme Fusion Proteins Facilitate G Protein-Coupled Receptor Crystallogenesis. Structure. 2014, 22, 1657. (PMID: 25450769) | |
 | Rattus norvegicus (Rat) (P08483) | M3-mT4L receptor bound to tiotropium PDB File: | X-Ray Diffraction | 2.8 | 2014-11-26 | Thorsen et al. Modified T4 Lysozyme Fusion Proteins Facilitate G Protein-Coupled Receptor Crystallogenesis. Structure. 2014, 22, 1657. (PMID: 25450769) | |
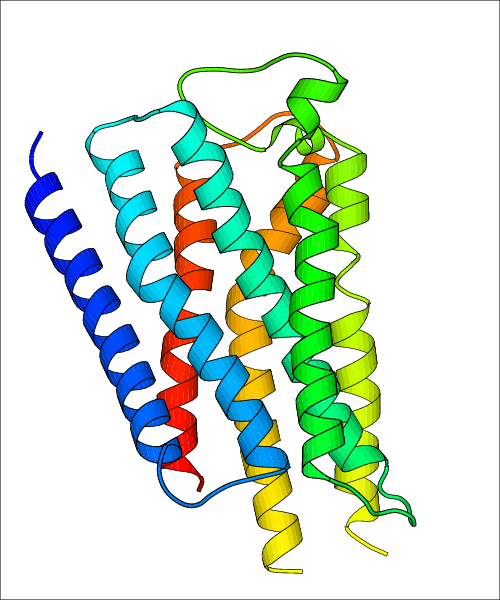 | Rattus norvegicus (Rat) (P08483) | M3-mT4L receptor bound to NMS PDB File: | X-Ray Diffraction | 3.7 | 2014-11-26 | Thorsen et al. Modified T4 Lysozyme Fusion Proteins Facilitate G Protein-Coupled Receptor Crystallogenesis. Structure. 2014, 22, 1657. (PMID: 25450769) | |
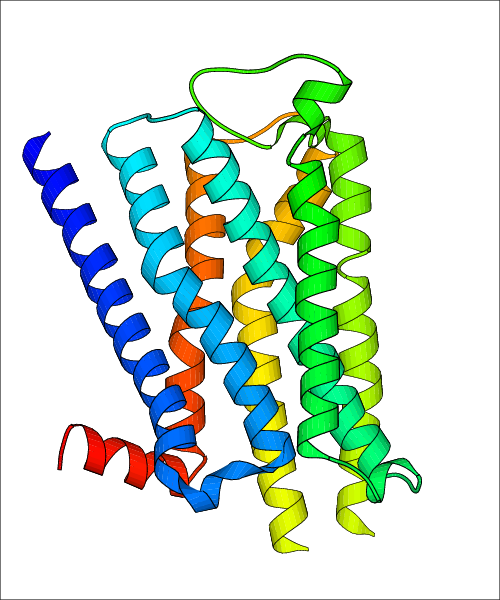 | Rattus norvegicus (Rat) (P08483) | M3 muscarinic acetylcholine receptor in complex with a selective antagonist PDB File: | X-Ray Diffraction | 3.1 | 2018-11-28 | Liu et al. Structure-guided development of selective M3 muscarinic acetylcholine receptor antagonists. Proc. Natl. Acad. Sci. U.S.A. 2018, 115, 12046. (PMID: 30404914) |
| PDB ID | Species (UniProt ID) | Description | Method | Resolution (Å) | Released | Reference | |
|---|---|---|---|---|---|---|---|
 | Homo sapiens (Human) (P08173) | Structure of the M4 muscarinic acetylcholine receptor (M4-mT4L) bound to tiotropium PDB File: | X-Ray Diffraction | 2.6 | 2016-03-16 | Thal et al. Crystal structures of the M1 and M4 muscarinic acetylcholine receptors. Nature. 2016, 531, 335. (PMID: 26958838) |
| PDB ID | Species (UniProt ID) | Description | Method | Resolution (Å) | Released | Reference | |
|---|---|---|---|---|---|---|---|
 | Homo sapiens (Human) (P08912) | Structure of the M5 muscarinic acetylcholine receptor (M5-T4L) bound to tiotropium PDB File: | X-Ray Diffraction | 2.54 | 2019-12-11 | Vuckovic et al. Crystal structure of the M5muscarinic acetylcholine receptor. Proc.Natl.Acad.Sci.USA. 2019. (PMID: 31772027) |
| PDB ID | Species (UniProt ID) | Description | Method | Resolution (Å) | Released | Reference | |
|---|---|---|---|---|---|---|---|
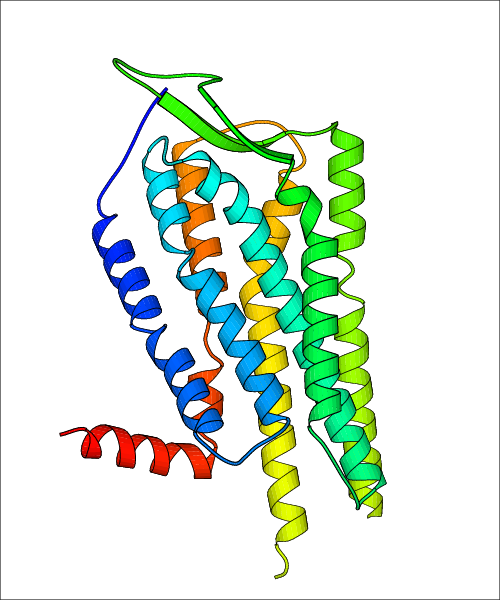 | Homo sapiens (Human) (P25929) | The Crystal Structure of Human Neuropeptide Y Y1 Receptor with BMS-193885 PDB File: | X-Ray Diffraction | 3.0 | 2018-04-25 | Yang et al. Structural basis of ligand binding modes at the neuropeptide Y Y1receptor. Nature. 2018, 556, 520. (PMID: 29670288) | |
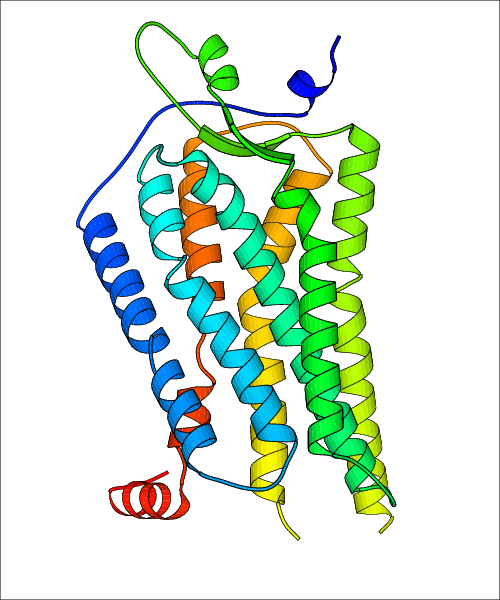 | Homo sapiens (Human) (P25929) | The Crystal Structure of human neuropeptide Y Y1 receptor with UR-MK299 PDB File: | X-Ray Diffraction | 2.7 | 2018-04-25 | Yang et al. Structural basis of ligand binding modes at the neuropeptide Y Y1receptor. Nature. 2018, 556, 520. (PMID: 29670288) |
| PDB ID | Species (UniProt ID) | Description | Method | Resolution (Å) | Released | Reference | |
|---|---|---|---|---|---|---|---|
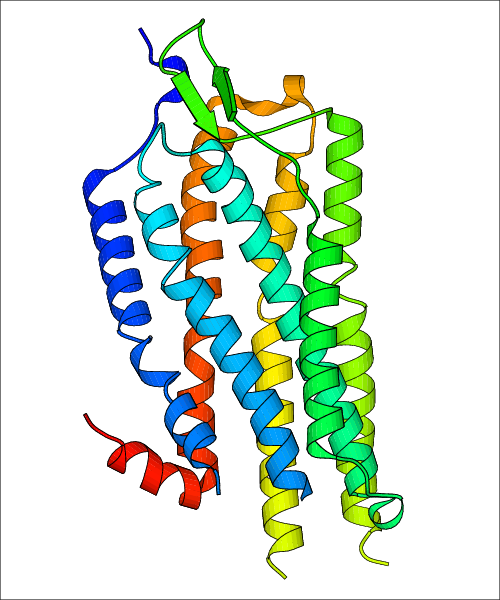 | Rattus norvegicus (Rat) (P20789) | Structure of Thermostable Agonist-bound Neurotensin Receptor 1 Mutant without Lysozyme Fusion PDB File: | X-Ray Diffraction | 3.0 | 2014-01-29 | Egloff et al. Structure of Signaling-Competent Neurotensin Receptor 1 Obtained by Directed Evolution in Escherichia Coli. Proc.Natl.Acad.Sci.USA. 2014, 111, E655. (PMID: 24453215) | |
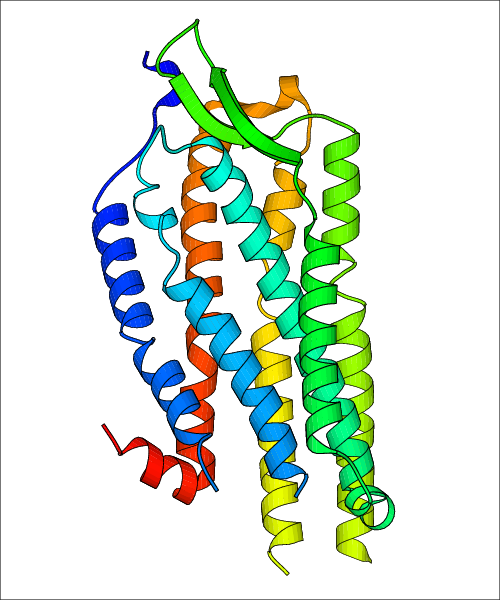 | Rattus norvegicus (Rat) (P20789) | High Resolution Structure of Thermostable Agonist-bound Neurotensin Receptor 1 Mutant without Lysozyme Fusion PDB File: | X-Ray Diffraction | 2.75 | 2014-01-29 | Egloff et al. Structure of Signaling-Competent Neurotensin Receptor 1 Obtained by Directed Evolution in Escherichia Coli. Proc.Natl.Acad.Sci.USA. 2014, 111, E655. (PMID: 24453215) | |
 | Rattus norvegicus (Rat) (P20789) | High Resolution Structure of Evolved Agonist-bound Neurotensin Receptor 1 Mutant without Lysozyme Fusion PDB File: | X-Ray Diffraction | 3.1 | 2014-01-29 | Egloff et al. Structure of Signaling-Competent Neurotensin Receptor 1 Obtained by Directed Evolution in Escherichia Coli. Proc.Natl.Acad.Sci.USA. 2014, 111, E655. (PMID: 24453215) | |
 | Rattus norvegicus (Rat) (P20789) | Structure of Evolved Agonist-bound Neurotensin Receptor 1 Mutant without Lysozyme Fusion PDB File: | X-Ray Diffraction | 3.57 | 2014-01-29 | Egloff et al. Structure of Signaling-Competent Neurotensin Receptor 1 Obtained by Directed Evolution in Escherichia Coli. Proc.Natl.Acad.Sci.USA. 2014, 111, E655. (PMID: 24453215) | |
 | Rattus norvegicus (Rat) (P20789) | The crystal structure of the neurotensin receptor NTS1 in complex with neurotensin (8-13) PDB File: | X-Ray Diffraction | 2.8 | 2012-10-17 | White et al. Structure of the agonist-bound neurotensin receptor. Nature. 2012, 490, 508. (PMID: 23051748) | |
 | Rattus norvegicus (Rat) (P20789) | Structure of active-like neurotensin receptor PDB File: | X-Ray Diffraction | 2.9 | 2015-07-29 | Krumm et al. Structural prerequisites for G-protein activation by the neurotensin receptor. Nat Commun. 2015, 6, 7895. (PMID: 26205105) | |
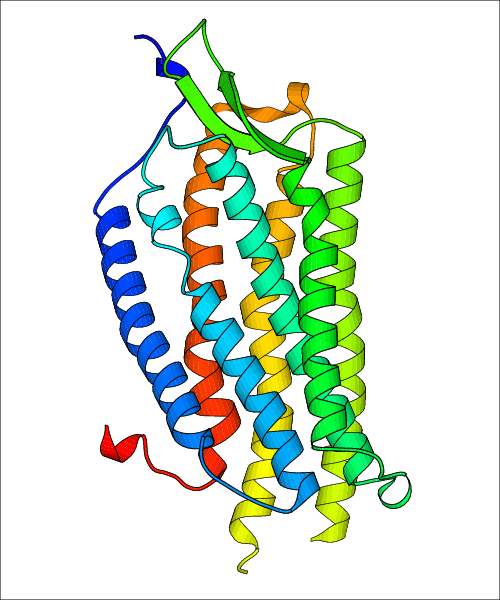 | Rattus norvegicus (Rat) (P20789) | Structure of active-like neurotensin receptor PDB File: | X-Ray Diffraction | 2.6 | 2015-07-29 | Krumm et al. Structural prerequisites for G-protein activation by the neurotensin receptor. Nat Commun. 2015, 6, 7895. (PMID: 26205105) | |
 | Rattus norvegicus (Rat) (P20789) | STRUCTURE OF CONSTITUTIVELY ACTIVE NEUROTENSIN RECEPTOR PDB File: | X-Ray Diffraction | 3.3 | 2016-12-21 | Krumm et al. Structure and dynamics of a constitutively active neurotensin receptor. Sci Rep. 2016, 6, 38564. (PMID: 27924846) | |
 | Homo sapiens (Human) (P30989) | human Neurotensin Receptor 1 (hNTSR1) - Gi1 Protein Complex in canonical conformation (C state) PDB File: | Electron Microscopy | 3.0 | 2019-07-10 | Kato et al. Conformational transitions of a neurotensin receptor 1-Gi1complex. Nature. 2019. (PMID: 31243364) | |
 | Homo sapiens (Human) (P30989) | human Neurotensin Receptor 1 (hNTSR1) - Gi1 Protein Complex in non-canonical conformation (NC state) PDB File: | Electron Microscopy | 3.0 | 2019-07-10 | Kato et al. Conformational transitions of a neurotensin receptor 1-Gi1complex. Nature. 2019. (PMID: 31243364) | |
 | Homo sapiens (Human) (P30989) | A complex structure of arrestin-2 bound to neurotensin receptor 1 PDB File: | Electron Microscopy | 4.9 | 2019-12-04 | Yin et al. A complex structure of arrestin-2 bound to a G protein-coupled receptor. Cell Res. 2019, 29, 971. (PMID: 31776446) |
| PDB ID | Species (UniProt ID) | Description | Method | Resolution (Å) | Released | Reference | |
|---|---|---|---|---|---|---|---|
 | Homo sapiens (Human) (P41146) | Structure of the N/OFQ Opioid Receptor in Complex with a Peptide Mimetic PDB File: | X-Ray Diffraction | 3.01 | 2012-04-25 | Thompson et al. Structure of the nociceptin/orphanin FQ receptor in complex with a peptide mimetic. Nature. 2012, 485, 395. (PMID: 22596163) | |
 | Homo sapiens (Human) (P41146) | The crystal structure of nociceptin/orphanin FQ peptide receptor (NOP) in complex with C-35 (PSI Community Target) PDB File: | X-Ray Diffraction | 3.0 | 2015-10-21 | Miller et al. The Importance of Ligand-Receptor Conformational Pairs in Stabilization: Spotlight on the N/OFQ G Protein-Coupled Receptor. Structure. 2015, 23, 2291. (PMID: 26526853) | |
 | Homo sapiens (Human) (P41146) | The crystal structure of nociceptin/orphanin FQ peptide receptor (NOP) in complex with SB-612111 (PSI Community Target) PDB File: | X-Ray Diffraction | 3.0 | 2015-10-21 | Miller et al. The Importance of Ligand-Receptor Conformational Pairs in Stabilization: Spotlight on the N/OFQ G Protein-Coupled Receptor. Structure. 2015, 23, 2291. (PMID: 26526853) |
| PDB ID | Species (UniProt ID) | Description | Method | Resolution (Å) | Released | Reference | |
|---|---|---|---|---|---|---|---|
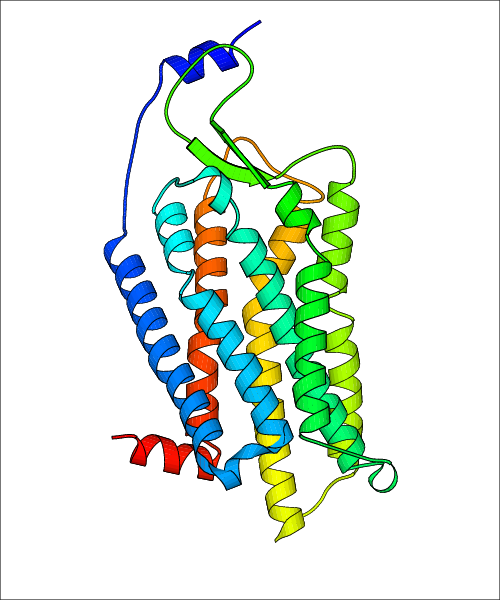 | Homo sapiens (Human) (O43613) | Structures of the human OX1 orexin receptor bound to selective and dual antagonists PDB File: | X-Ray Diffraction | 2.75 | 2016-03-09 | Yin et al. Structure and ligand-binding mechanism of the human OX1 and OX2 orexin receptors. Nat.Struct.Mol.Biol. 2016, 23, 293. (PMID: 26950369) | |
 | Homo sapiens (Human) (O43613) | Structures of the human OX1 orexin receptor bound to selective and dual antagonists PDB File: | X-Ray Diffraction | 2.83 | 2016-03-09 | Yin et al. Structure and ligand-binding mechanism of the human OX1 and OX2 orexin receptors. Nat.Struct.Mol.Biol. 2016, 23, 293. (PMID: 26950369) | |
 | Homo sapiens (Human) (O43613) | Crystal structure of the Orexin-1 receptor in complex with suvorexant at 2.29 A resolution PDB File: | X-Ray Diffraction | 2.26 | 2020-01-01 | Rappas et al. Comparison of orexin 1 and orexin 2 ligand binding modes using X-ray crystallography and computational analysis. J.Med.Chem. 2019. (PMID: 31860301) | |
 | Homo sapiens (Human) (O43613) | Crystal structure of the Orexin-1 receptor in complex with EMPA PDB File: | X-Ray Diffraction | 2.11 | 2020-01-01 | Rappas et al. Comparison of orexin 1 and orexin 2 ligand binding modes using X-ray crystallography and computational analysis. J.Med.Chem. 2019. (PMID: 31860301) | |
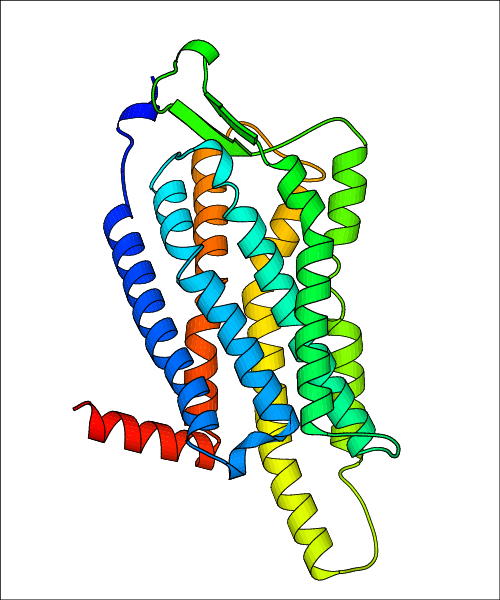 | Homo sapiens (Human) (O43613) | Crystal structure of the Orexin-1 receptor in complex with daridorexant PDB File: | X-Ray Diffraction | 3.04 | 2020-01-01 | Rappas et al. Comparison of orexin 1 and orexin 2 ligand binding modes using X-ray crystallography and computational analysis. J.Med.Chem. 2019. (PMID: 31860301) | |
 | Homo sapiens (Human) (O43613) | Crystal structure of the Orexin-1 receptor in complex with ACT-462206 PDB File: | X-Ray Diffraction | 3.01 | 2020-01-01 | Rappas et al. Comparison of orexin 1 and orexin 2 ligand binding modes using X-ray crystallography and computational analysis. J.Med.Chem. 2019. (PMID: 31860301) | |
 | Homo sapiens (Human) (O43613) | Crystal structure of the Orexin-1 receptor in complex with filorexant PDB File: | X-Ray Diffraction | 2.34 | 2020-01-01 | Rappas et al. Comparison of orexin 1 and orexin 2 ligand binding modes using X-ray crystallography and computational analysis. J.Med.Chem. 2019. (PMID: 31860301) | |
 | Homo sapiens (Human) (O43613) | Crystal structure of the Orexin-1 receptor in complex with Compound 16 PDB File: | X-Ray Diffraction | 2.3 | 2020-01-01 | Rappas et al. Comparison of orexin 1 and orexin 2 ligand binding modes using X-ray crystallography and computational analysis. J.Med.Chem. 2019. (PMID: 31860301) | |
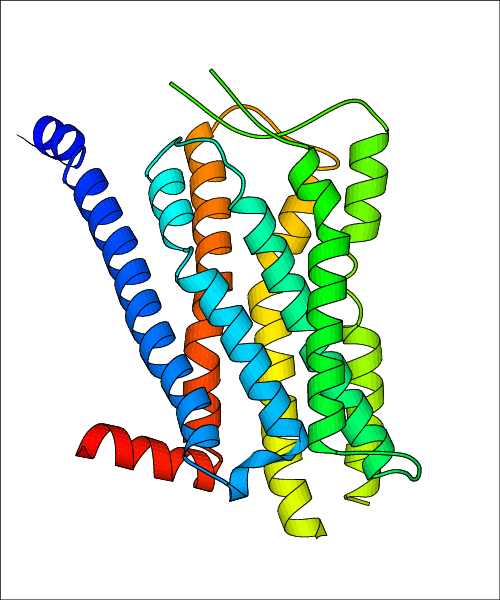 | Homo sapiens (Human) (O43613) | Crystal structure of the Orexin-1 receptor in complex with Compound 14 PDB File: | X-Ray Diffraction | 2.55 | 2020-01-01 | Rappas et al. Comparison of orexin 1 and orexin 2 ligand binding modes using X-ray crystallography and computational analysis. J.Med.Chem. 2019. (PMID: 31860301) | |
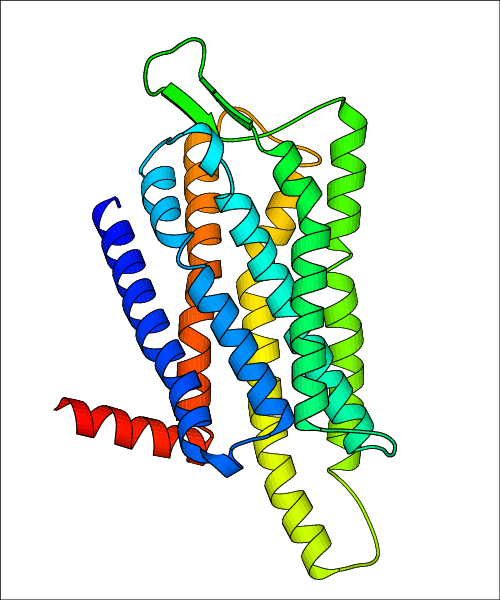 | Homo sapiens (Human) (O43613) | Crystal structure of the Orexin-1 receptor in complex with SB-334867 PDB File: | X-Ray Diffraction | 2.66 | 2020-01-01 | Rappas et al. Comparison of orexin 1 and orexin 2 ligand binding modes using X-ray crystallography and computational analysis. J.Med.Chem. 2019. (PMID: 31860301) | |
 | Homo sapiens (Human) (O43613) | Crystal structure of the Orexin-1 receptor in complex with SB-408124 PDB File: | X-Ray Diffraction | 2.65 | 2020-01-01 | Rappas et al. Comparison of orexin 1 and orexin 2 ligand binding modes using X-ray crystallography and computational analysis. J.Med.Chem. 2019. (PMID: 31860301) |
| PDB ID | Species (UniProt ID) | Description | Method | Resolution (Å) | Released | Reference | |
|---|---|---|---|---|---|---|---|
 | Homo sapiens (Human) (O43614) | Crystal structure of the human OX2 orexin receptor bound to the insomnia drug Suvorexant PDB File: | X-Ray Diffraction | 2.5 | 2015-01-14 | Yin et al. Crystal structure of the human OX2 orexin receptor bound to the insomnia drug suvorexant. Nature. 2015, 519, 247. (PMID: 25533960) | |
 | Homo sapiens (Human) (O43614) | Crystal structure of human orexin 2 receptor bound to the selective antagonist EMPA determined by the synchrotron light source at SPring-8. PDB File: | X-Ray Diffraction | 1.96 | 2017-11-29 | Suno et al. Crystal Structures of Human Orexin 2 Receptor Bound to the Subtype-Selective Antagonist EMPA. Structure. 2018, 26, 7. (PMID: 29225076) | |
 | Homo sapiens (Human) (O43614) | Crystal structures of human orexin 2 receptor bound to the selective antagonist EMPA determined by serial femtosecond crystallography at SACLA PDB File: | X-Ray Diffraction | 2.3 | 2017-12-13 | Suno et al. Crystal Structures of Human Orexin 2 Receptor Bound to the Subtype-Selective Antagonist EMPA. Structure. 2018, 26, 7. (PMID: 29225076) | |
 | Homo sapiens (Human) (O43614) | Crystal structure of the Orexin-2 receptor in complex with EMPA at 2.74 A resolution PDB File: | X-Ray Diffraction | 2.74 | 2020-01-01 | Rappas et al. Comparison of orexin 1 and orexin 2 ligand binding modes using X-ray crystallography and computational analysis. J.Med.Chem. 2019. (PMID: 31860301) | |
 | Homo sapiens (Human) (O43614) | Crystal structure of the Orexin-2 receptor in complex with suvorexant at 2.76 A resolution PDB File: | X-Ray Diffraction | 2.74 | 2020-01-01 | Rappas et al. Comparison of orexin 1 and orexin 2 ligand binding modes using X-ray crystallography and computational analysis. J.Med.Chem. 2019. (PMID: 31860301) | |
 | Homo sapiens (Human) (O43614) | Crystal structure of the Orexin-2 receptor in complex with HTL6641 at 2.61 A resolution PDB File: | X-Ray Diffraction | 2.61 | 2020-01-01 | Rappas et al. Comparison of orexin 1 and orexin 2 ligand binding modes using X-ray crystallography and computational analysis. J.Med.Chem. 2019. (PMID: 31860301) |
| PDB ID | Species (UniProt ID) | Description | Method | Resolution (Å) | Released | Reference | |
|---|---|---|---|---|---|---|---|
 | Homo sapiens (Human) (P47900) | The human P2Y1 receptor in complex with BPTU PDB File: | X-Ray Diffraction | 2.2 | 2015-04-01 | Zhang et al. Two disparate ligand-binding sites in the human P2Y1 receptor. Nature. 2015, 520, 317. (PMID: 25822790) | |
 | Homo sapiens (Human) (P47900) | The human P2Y1 receptor in complex with MRS2500 PDB File: | X-Ray Diffraction | 2.7 | 2015-04-01 | Zhang et al. Two disparate ligand-binding sites in the human P2Y1 receptor. Nature. 2015, 520, 317. (PMID: 25822790) |
| PDB ID | Species (UniProt ID) | Description | Method | Resolution (Å) | Released | Reference | |
|---|---|---|---|---|---|---|---|
 | Homo sapiens (Human) (Q9H244) | Structure of the human P2Y12 receptor in complex with an antithrombotic drug PDB File: | X-Ray Diffraction | 2.62 | 2014-03-26 | Zhang et al. Structure of the human P2Y12 receptor in complex with an antithrombotic drug. Nature. 2014, 509, 115. (PMID: 24670650) | |
 | Homo sapiens (Human) (Q9H244) | Crystal structure of P2Y12 receptor in complex with 2MeSADP PDB File: | X-Ray Diffraction | 2.5 | 2014-04-30 | Zhang et al. Agonist-bound structure of the human P2Y12 receptor. Nature. 2014, 509, 119. (PMID: 24784220) | |
 | Homo sapiens (Human) (Q9H244) | Crystal structure of P2Y12 receptor in complex with 2MeSATP PDB File: | X-Ray Diffraction | 3.1 | 2014-04-30 | Zhang et al. Agonist-bound structure of the human P2Y12 receptor. Nature. 2014, 509, 119. (PMID: 24784220) |
| PDB ID | Species (UniProt ID) | Description | Method | Resolution (Å) | Released | Reference | |
|---|---|---|---|---|---|---|---|
 | Homo sapiens (Human) (P25105) | Crystal structure of the human platelet-activating factor receptor in complex with SR 27417 PDB File: | X-Ray Diffraction | 2.81 | 2018-06-20 | Cao et al. Structural basis for signal recognition and transduction by platelet-activating-factor receptor. Nat. Struct. Mol. Biol. 2018, 25, 488. (PMID: 29808000) | |
 | Homo sapiens (Human) (P25105) | Crystal structure of the human platelet-activating factor receptor in complex with ABT-491 PDB File: | X-Ray Diffraction | 2.9 | 2018-06-20 | Cao et al. Structural basis for signal recognition and transduction by platelet-activating-factor receptor. Nat. Struct. Mol. Biol. 2018, 25, 488. (PMID: 29808000) |
| PDB ID | Species (UniProt ID) | Description | Method | Resolution (Å) | Released | Reference | |
|---|---|---|---|---|---|---|---|
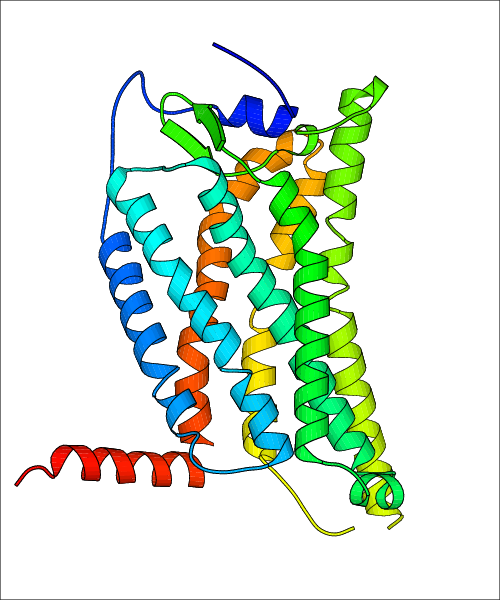 | Homo sapiens (Human) (Q9Y5Y4) | Crystal structure of the prostaglandin D2 receptor CRTH2 with fevipiprant PDB File: | X-Ray Diffraction | 2.8 | 2018-10-03 | Wang et al. Structures of the Human PGD2Receptor CRTH2 Reveal Novel Mechanisms for Ligand Recognition. Mol. Cell. 2018, 72, 48. (PMID: 30220562) | |
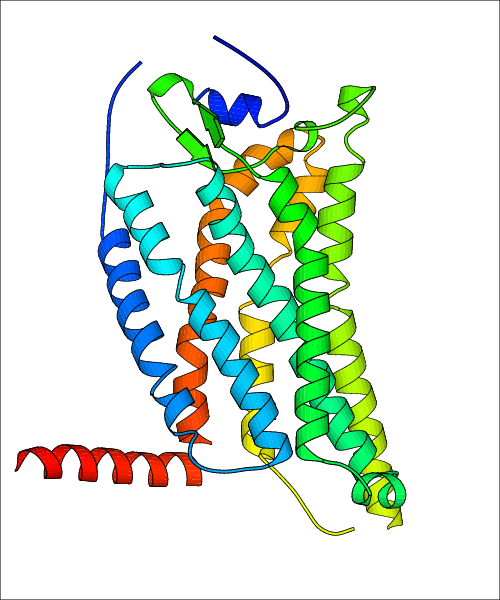 | Homo sapiens (Human) (Q9Y5Y4) | Crystal structure of the prostaglandin D2 receptor CRTH2 with CAY10471 PDB File: | X-Ray Diffraction | 2.74 | 2018-10-03 | Wang et al. Structures of the Human PGD2Receptor CRTH2 Reveal Novel Mechanisms for Ligand Recognition. Mol. Cell. 2018, 72, 48. (PMID: 30220562) |
| PDB ID | Species (UniProt ID) | Description | Method | Resolution (Å) | Released | Reference | |
|---|---|---|---|---|---|---|---|
 | Homo sapiens (Human) (P43115) | Crystal structure of the human prostaglandin E receptor EP3 bound to prostaglandin E2 PDB File: | X-Ray Diffraction | 2.9 | 2018-12-05 | Toyoda et al. Ligand binding to human prostaglandin E receptor EP4at the lipid-bilayer interface. Nat. Chem. Biol. 2019, 15, 18. (PMID: 30510193) | |
 | Homo sapiens (Human) (P43115) | Crystal structure of EP3 receptor bound to misoprostol-FA PDB File: | X-Ray Diffraction | 2.5 | 2018-12-05 | Audet et al. Crystal structure of misoprostol bound to the labor inducer prostaglandin E2receptor. Nat. Chem. Biol. 2019, 15, 11. (PMID: 30510194) |
| PDB ID | Species (UniProt ID) | Description | Method | Resolution (Å) | Released | Reference | |
|---|---|---|---|---|---|---|---|
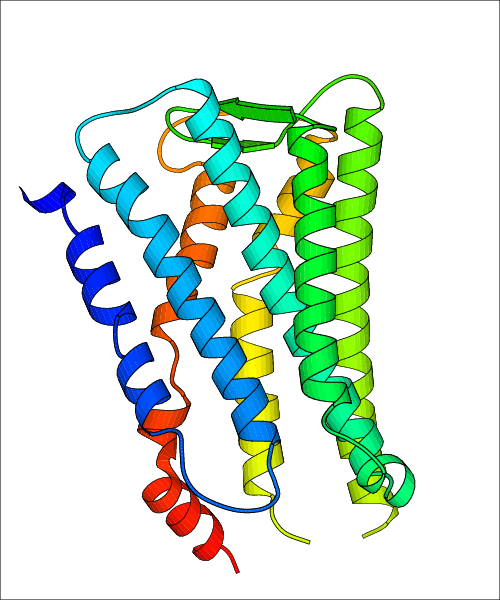 | Homo sapiens (Human) (P35408) | Anomalous bromine signal of the human prostaglandin E receptor EP4 in complex with Fab and ONO-AE3-208-Br derivative PDB File: | X-Ray Diffraction | 4.2 | 2018-12-05 | Toyoda et al. Ligand binding to human prostaglandin E receptor EP4at the lipid-bilayer interface. Nat. Chem. Biol. 2019, 15, 18. (PMID: 30510193) | |
 | Homo sapiens (Human) (P35408) | Crystal structure of the human prostaglandin E receptor EP4 in complex with Fab and ONO-AE3-208 PDB File: | X-Ray Diffraction | 3.2 | 2018-12-05 | Toyoda et al. Ligand binding to human prostaglandin E receptor EP4at the lipid-bilayer interface. Nat. Chem. Biol. 2019, 15, 18. (PMID: 30510193) |
| PDB ID | Species (UniProt ID) | Description | Method | Resolution (Å) | Released | Reference | |
|---|---|---|---|---|---|---|---|
 | Homo sapiens (Human) (P25116) | Crystal structure of human protease-activated receptor 1 (PAR1) bound with antagonist vorapaxar at 2.2 angstrom PDB File: | X-Ray Diffraction | 2.2 | 2012-12-12 | Zhang et al. High-resolution crystal structure of human protease-activated receptor 1. Nature. 2012, 492, 387. (PMID: 23222541) |
| PDB ID | Species (UniProt ID) | Description | Method | Resolution (Å) | Released | Reference | |
|---|---|---|---|---|---|---|---|
 | Homo sapiens (Human) (P55085) | Crystal structure of a thermostabilised human protease-activated receptor-2 (PAR2) in complex with AZ8838 at 2.8 angstrom resolution PDB File: | X-Ray Diffraction | 2.8 | 2017-05-03 | Cheng et al. Structural insight into allosteric modulation of protease-activated receptor 2. Nature. 2017, 545, 112. (PMID: 28445455) | |
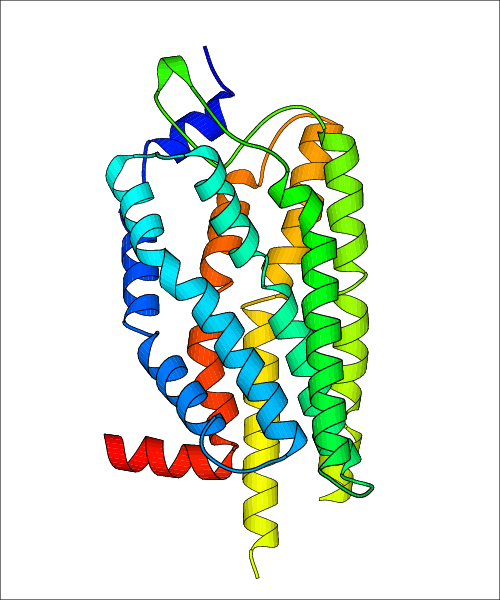 | Homo sapiens (Human) (P55085) | Crystal structure of a thermostabilised human protease-activated receptor-2 (PAR2) in complex with AZ3451 at 3.6 angstrom resolution PDB File: | X-Ray Diffraction | 3.6 | 2017-05-03 | Cheng et al. Structural insight into allosteric modulation of protease-activated receptor 2. Nature. 2017, 545, 112. (PMID: 28445455) | |
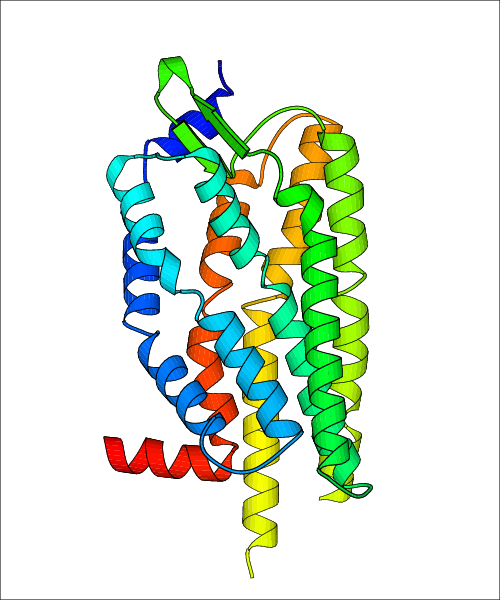 | Homo sapiens (Human) (P55085) | Crystal structure of a thermostabilised human protease-activated receptor-2 (PAR2) in ternary complex with Fab3949 and AZ7188 at 4.0 angstrom resolution PDB File: | X-Ray Diffraction | 4.0 | 2017-05-03 | Cheng et al. Structural insight into allosteric modulation of protease-activated receptor 2. Nature. 2017, 545, 112. (PMID: 28445455) |
| PDB ID | Species (UniProt ID) | Description | Method | Resolution (Å) | Released | Reference | |
|---|---|---|---|---|---|---|---|
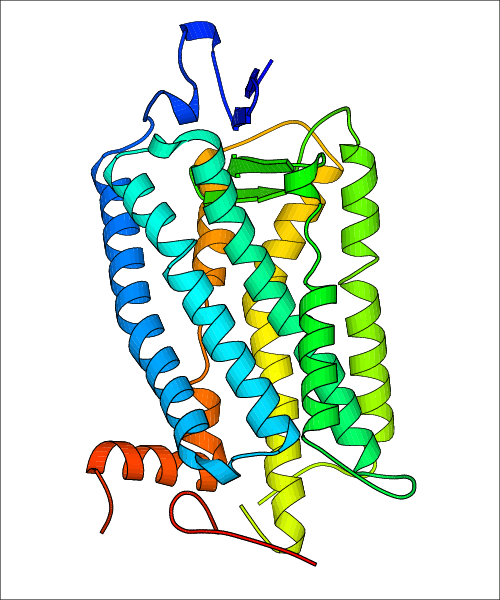 | Bos taurus (Bovine) (P02699) | CRYSTAL STRUCTURE OF BOVINE RHODOPSIN PDB File: | X-Ray Diffraction | 2.8 | 2000-08-04 | Palczewski et al. Crystal structure of rhodopsin: A G protein-coupled receptor. Science. 2000, 289, 739. (PMID: 10926528) | |
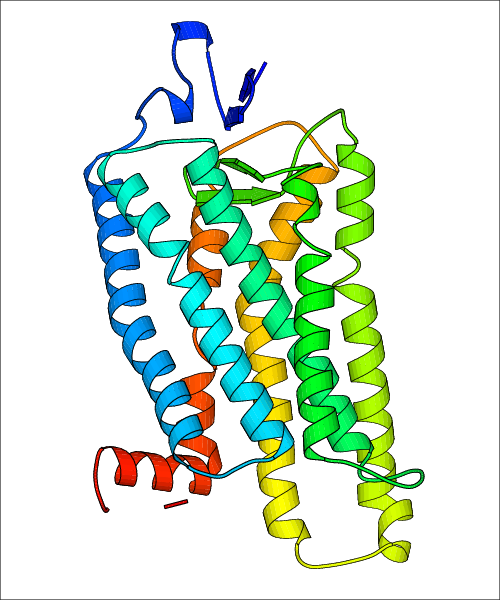 | Bos taurus (Bovine) (P02699) | Structure of Bovine Rhodopsin in a Trigonal Crystal Form PDB File: | X-Ray Diffraction | 2.65 | 2003-11-20 | Li et al. Structure of Bovine Rhodopsin in a Trigonal Crystal Form. J.Mol.Biol. 2004, 343, 1409. (PMID: 15491621) | |
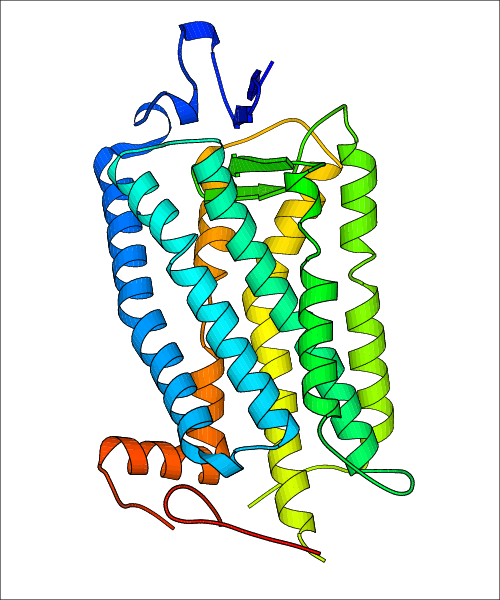 | Bos taurus (Bovine) (P02699) | CRYSTAL STRUCTURE OF BOVINE RHODOPSIN PDB File: | X-Ray Diffraction | 2.8 | 2001-07-04 | Teller et al. Advances in determination of a high-resolution three-dimensional structure of rhodopsin, a model of G-protein-coupled receptors (GPCRs). Biochemistry. 2001, 40, 7761. (PMID: 11425302) | |
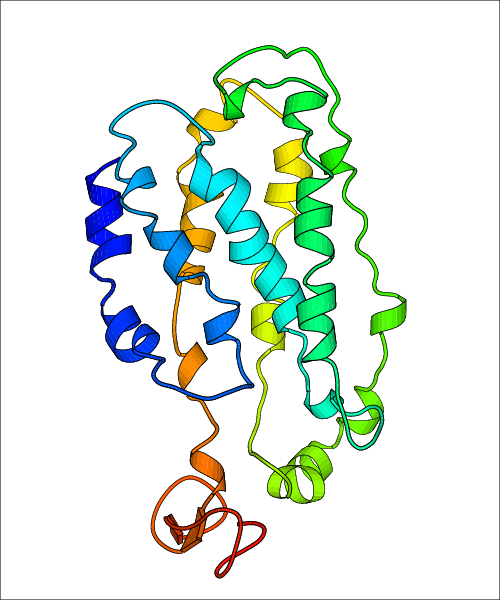 | Bos taurus (Bovine) (P02699) | Structure of bovine rhodopsin (dark adapted) PDB File: | Solution NMR | N/A | 2001-10-05 | Yeagle et al. Studies on the structure of the G-protein-coupled receptor rhodopsin including the putative G-protein binding site in unactivated and activated forms. Biochemistry. 2001, 40, 11932. (PMID: 11570894) | |
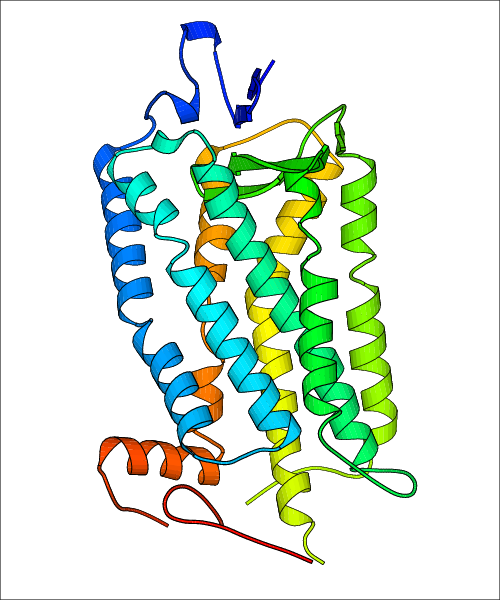 | Bos taurus (Bovine) (P02699) | Crystal structure of bovine rhodopsin at 2.6 angstroms RESOLUTION PDB File: | X-Ray Diffraction | 2.6 | 2002-05-15 | Okada et al. Functional role of internal water molecules in rhodopsin revealed by X-ray crystallography. Proc.Natl.Acad.Sci.USA. 2002, 99, 5982. (PMID: 11972040) | |
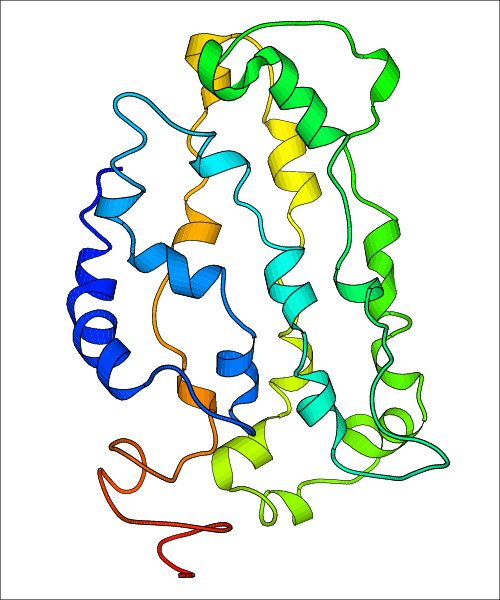 | Bos taurus (Bovine) (P02699) | STRUCTURE OF BOVINE RHODOPSIN (Metarhodopsin II) PDB File: | Solution NMR | N/A | 2002-07-10 | Choi et al. Structural studies of metarhodopsin II, the activated form of the G-protein coupled receptor, rhodopsin. Biochemistry. 2002, 41, 7318. (PMID: 12044163) | |
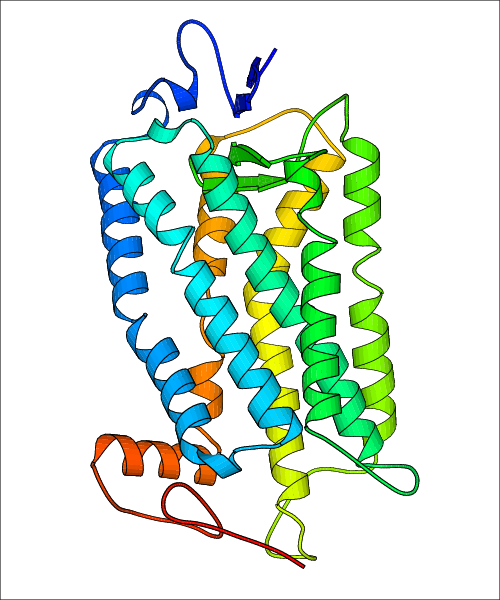 | Bos taurus (Bovine) (P02699) | Crystal Structure of Bovine Rhodopsin at 2.2 Angstroms Resolution PDB File: | X-Ray Diffraction | 2.2 | 2004-10-12 | Okada et al. The retinal conformation and its environment in rhodopsin in light of a new 22 A crystal structure. J.Mol.Biol. 2004, 342, 571. (PMID: 15327956) | |
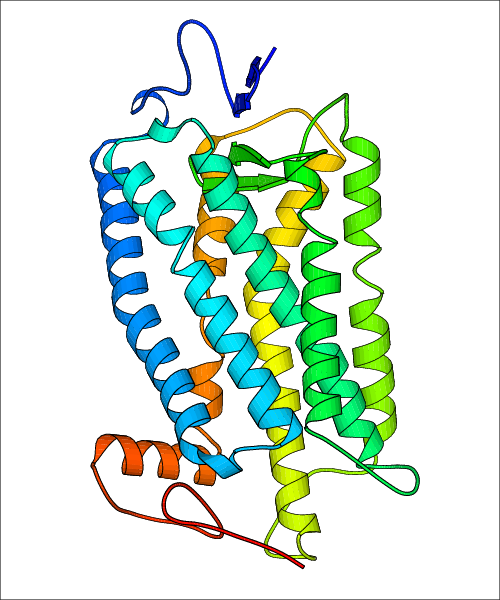 | Bos taurus (Bovine) (P02699) | Crystallographic model of bathorhodopsin PDB File: | X-Ray Diffraction | 2.6 | 2006-09-02 | Nakamichi et al. Crystallographic analysis of primary visual photochemistry. Angew.Chem.Int.Ed.Engl. 2006, 45, 4270. (PMID: 16586416) | |
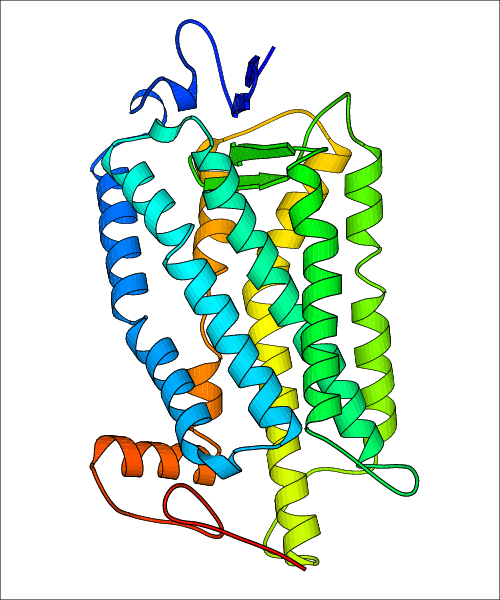 | Bos taurus (Bovine) (P02699) | Crystallographic model of lumirhodopsin PDB File: | X-Ray Diffraction | 2.8 | 2006-08-22 | Nakamichi et al. Local peptide movement in the photoreaction intermediate of rhodopsin. Proc.Natl.Acad.Sci.Usa. 2006, 103, 12729. (PMID: 16908857) | |
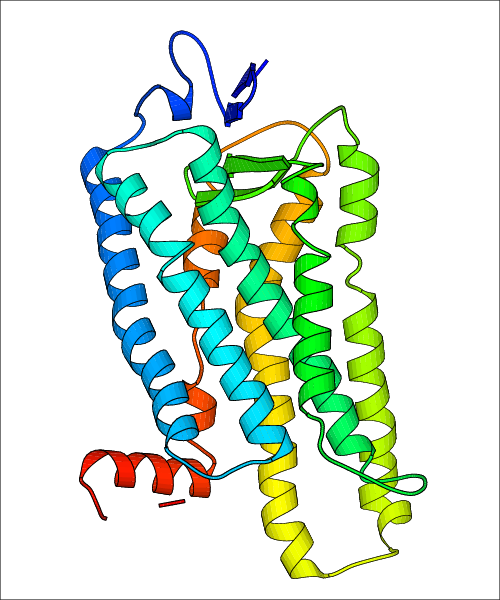 | Bos taurus (Bovine) (P02699) | Crystal structure of rhombohedral crystal form of ground-state rhodopsin PDB File: | X-Ray Diffraction | 3.8 | 2006-10-17 | Salom et al. Crystal structure of a photoactivated deprotonated intermediate of rhodopsin. Proc.Natl.Acad.Sci.Usa. 2006, 103, 16123. (PMID: 17060607) | |
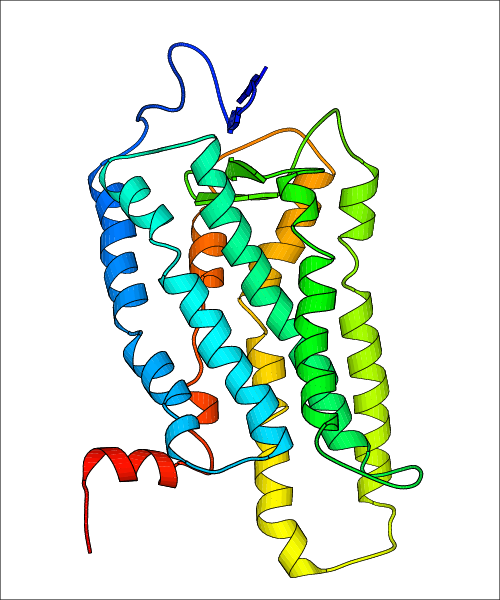 | Bos taurus (Bovine) (P02699) | Crystal structure of trigonal crystal form of ground-state rhodopsin PDB File: | X-Ray Diffraction | 4.1 | 2006-10-17 | Salom et al. Crystal structure of a photoactivated deprotonated intermediate of rhodopsin. Proc.Natl.Acad.Sci.Usa. 2006, 103, 16123. (PMID: 17060607) | |
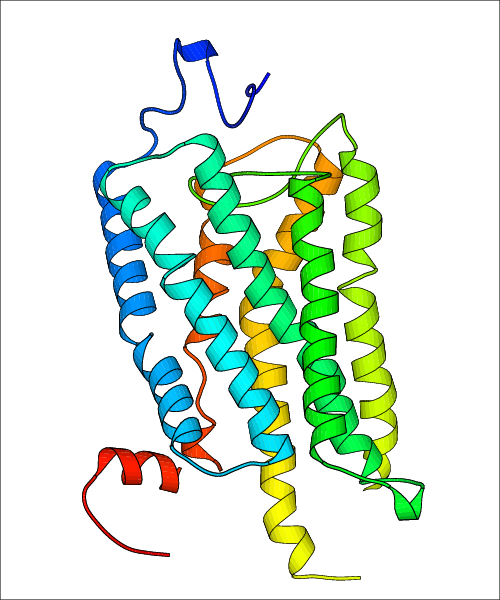 | Bos taurus (Bovine) (P02699) | Crystal structure of a photoactivated rhodopsin PDB File: | X-Ray Diffraction | 4.15 | 2006-10-17 | Salom et al. Crystal structure of a photoactivated deprotonated intermediate of rhodopsin. Proc.Natl.Acad.Sci.Usa. 2006, 103, 16123. (PMID: 17060607) | |
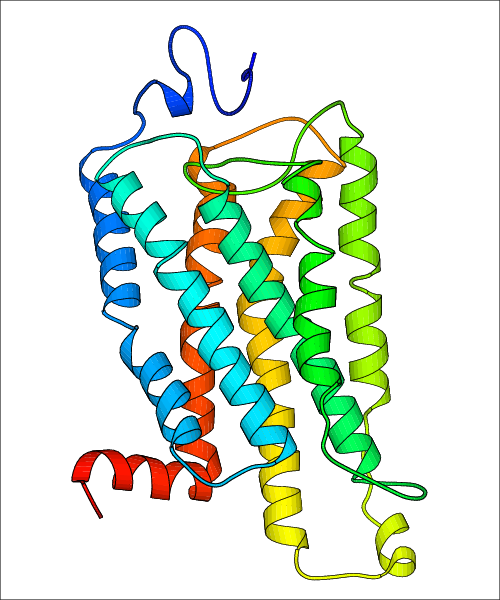 | Bos taurus (Bovine) (P02699) | Crystal structure of a rhodopsin stabilizing mutant expressed in mammalian cells PDB File: | X-Ray Diffraction | 3.4 | 2007-09-25 | Standfuss et al. Crystal Structure of a Thermally Stable Rhodopsin Mutant. J.Mol.Biol. 2007, 372, 1179. (PMID: 17825322) | |
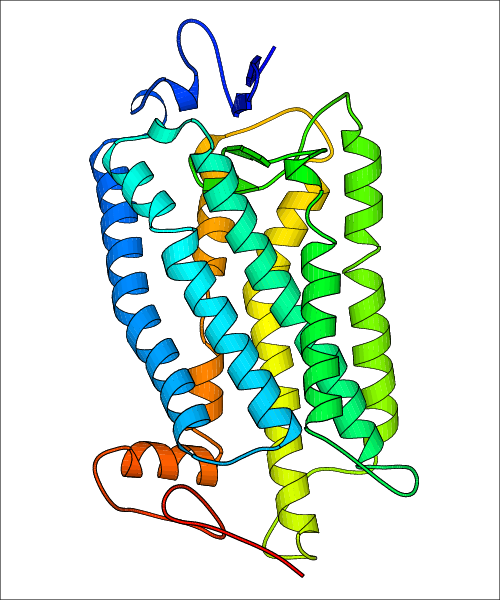 | Bos taurus (Bovine) (P02699) | Crystallographic model of 9-cis-rhodopsin PDB File: | X-Ray Diffraction | 2.95 | 2007-10-30 | Nakamichi et al. Photoisomerization mechanism of rhodopsin and 9-cis-rhodopsin revealed by x-ray crystallography. Biophys.J. 2007, 92, L106. (PMID: 17449675) | |
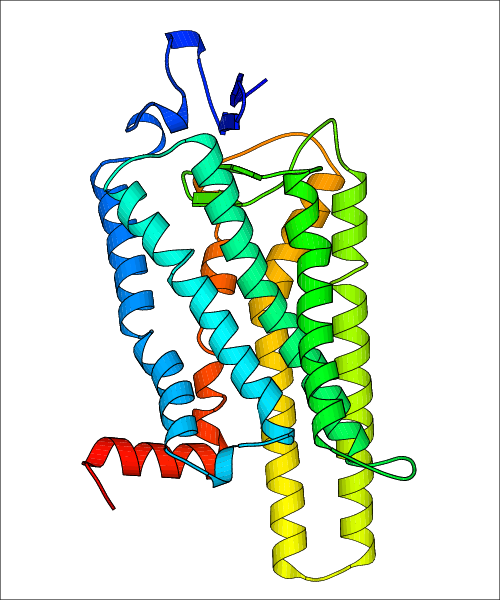 | Bos taurus (Bovine) (P02699) | CRYSTAL STRUCTURE OF THE CONSTITUTIVELY ACTIVE E113Q,D2C,D282C RHODOPSIN MUTANT WITH BOUND GALPHACT PEPTIDE. PDB File: | X-Ray Diffraction | 3.0 | 2011-03-16 | Standfuss et al. The Structural Basis of Agonist Induced Activation in Constitutively Active Rhodopsin. Nature. 2011, 471, 656. (PMID: 21389983) | |
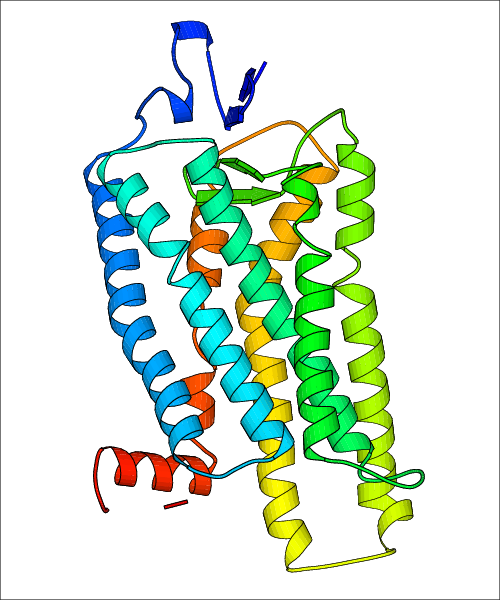 | Bos taurus (Bovine) (P02699) | Structure of ground-state bovine rhodospin in a hexagonal crystal form PDB File: | X-Ray Diffraction | 2.65 | 2008-08-05 | Stenkamp et al. Alternative models for two crystal structures of bovine rhodopsin. Acta Crystallogr.,Sect.D. 2008, 64, 902. (PMID: 18645239) | |
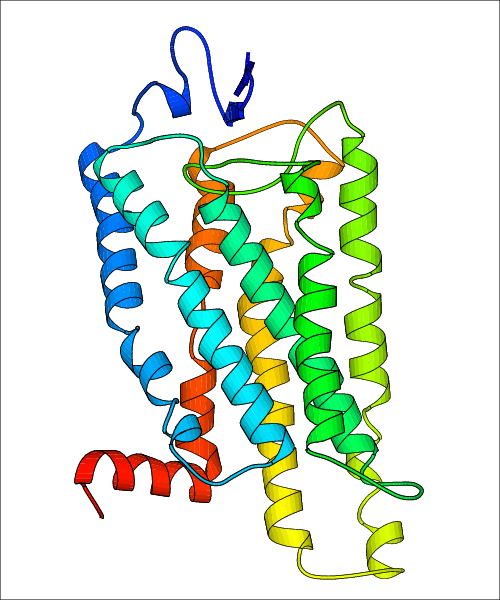 | Bos taurus (Bovine) (P02699) | Structure of a mutant bovine rhodopsin in hexagonal crystal form PDB File: | X-Ray Diffraction | 3.4 | 2008-08-05 | Stenkamp et al. Alternative models for two crystal structures of bovine rhodopsin. Acta Crystallogr.,Sect.D. 2008, 64, 902. (PMID: 18645239) | |
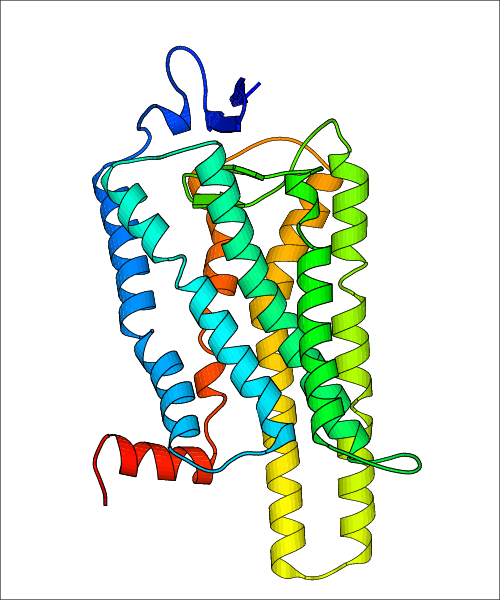 | Bos taurus (Bovine) (P02699) | Crystal Structure of Native Opsin: the G Protein-Coupled Receptor Rhodopsin in its Ligand-free State PDB File: | X-Ray Diffraction | 2.9 | 2008-06-24 | Park et al. Crystal structure of the ligand-free G-protein-coupled receptor opsin. Nature. 2008, 454, 183. (PMID: 18563085) | |
 | Bos taurus (Bovine) (P02699) | Crystal structure of the active G-protein-coupled receptor opsin in complex with a C-terminal peptide derived from the Galpha subunit of transducin PDB File: | X-Ray Diffraction | 3.2 | 2008-09-23 | Scheerer et al. Crystal structure of opsin in its G-protein-interacting conformation. Nature. 2008, 455, 497. (PMID: 18818650) | |
 | Bos taurus (Bovine) (P02699) | Crystal structure of bovine rhodopsin with beta-ionone PDB File: | X-Ray Diffraction | 2.6 | 2011-01-19 | Makino et al. Binding of more than one retinoid to visual opsins. Biophys.J. 2010, 99, 2366. (PMID: 20923672) | |
 | Bos taurus (Bovine) (P02699) | Crystal structure of Metarhodopsin II in complex with a C-terminal peptide derived from the Galpha subunit of transducin PDB File: | X-Ray Diffraction | 2.85 | 2011-03-09 | Choe et al. Crystal structure of metarhodopsin II. Nature. 2011, 471, 651. (PMID: 21389988) | |
 | Bos taurus (Bovine) (P02699) | Crystal structure of Metarhodopsin II PDB File: | X-Ray Diffraction | 3.0 | 2011-03-09 | Choe et al. Crystal structure of metarhodopsin II. Nature. 2011, 471, 651. (PMID: 21389988) | |
 | Bos taurus (Bovine) (P02699) | Crystal structure of the light-activated constitutively active N2C, M257Y,D282C rhodopsin mutant in complex with a peptide resembling the C-terminus of the Galpha-protein subunit (GaCT) PDB File: | X-Ray Diffraction | 3.3 | 2012-01-25 | Deupi et al. Stabilized G Protein Binding Site in the Structure of Constitutively Active Metarhodopsin-II. Proc.Natl.Acad.Sci.USA. 2012, 109, 119. (PMID: 22198838) | |
 | Bos taurus (Bovine) (P02699) | Night blindness causing G90D rhodopsin in complex with GaCT2 peptide PDB File: | X-Ray Diffraction | 2.9 | 2013-05-08 | Singhal et al. Insights Into Congenital Stationary Night Blindness Based on the Structure of G90D Rhodopsin. Embo Rep. 2013, 14, 520. (PMID: 23579341) | |
 | Bos taurus (Bovine) (P02699) | Night blindness causing G90D rhodopsin in the active conformation PDB File: | X-Ray Diffraction | 3.3 | 2013-04-24 | Singhal et al. Insights Into Congenital Stationary Night Blindness Based on the Structure of G90D Rhodopsin. Embo Rep. 2013, 14, 520. (PMID: 23579341) | |
 | Bos taurus (Bovine) (P02699) | Crystal structure of active conformation of GPCR opsin stabilized by octylglucoside PDB File: | X-Ray Diffraction | 2.65 | 2013-10-30 | Park et al. Opsin, a structural model for olfactory receptors?. Angew.Chem.Int.Ed.Engl. 2013, 52, 11021. (PMID: 24038729) | |
 | Bos taurus (Bovine) (P02699) | Crystal structure of the active G-protein-coupled receptor opsin in complex with the finger-loop peptide derived from the full-length arrestin-1 PDB File: | X-Ray Diffraction | 2.75 | 2014-09-17 | Szczepek et al. Crystal structure of a common GPCR-binding interface for G protein and arrestin. Nat Commun. 2014, 5, 4801. (PMID: 25205354) | |
 | Bos taurus (Bovine) (P02699) | Opsin/G(alpha) peptide complex stabilized by nonyl-glucoside PDB File: | X-Ray Diffraction | 2.29 | 2015-11-04 | Blankenship et al. The High-Resolution Structure of Activated Opsin Reveals a Conserved Solvent Network in the Transmembrane Region Essential for Activation. Structure. 2015, 23, 2358. (PMID: 26526852) | |
 | Bos taurus (Bovine) (P02699) | Crystal Structure of T94I rhodopsin mutant PDB File: | X-Ray Diffraction | 2.3 | 2016-08-10 | Singhal et al. Structural role of the T94I rhodopsin mutation in congenital stationary night blindness. Embo Rep. 2016, 17, 1431. (PMID: 27458239) | |
 | Bos taurus (Bovine) (P02699) | Crystal Structure of T94I rhodopsin mutant PDB File: | X-Ray Diffraction | 2.81 | 2016-08-10 | Singhal et al. Structural role of the T94I rhodopsin mutation in congenital stationary night blindness. Embo Rep. 2016, 17, 1431. (PMID: 27458239) | |
 | Bos taurus (Bovine) (P02699) | Crystal structure of Bos taurus opsin at 2.7 Angstrom PDB File: | X-Ray Diffraction | 2.7 | 2017-03-15 | Gulati et al. Photocyclic behavior of rhodopsin induced by an atypical isomerization mechanism. Proc. Natl. Acad. Sci. U.S.A. 2017, 114, E2608. (PMID: 28289214) | |
 | Bos taurus (Bovine) (P02699) | Crystal structure of Bos taurus opsin regenerated with 6-carbon ring retinal chromophore PDB File: | X-Ray Diffraction | 4.01 | 2017-03-15 | Gulati et al. Photocyclic behavior of rhodopsin induced by an atypical isomerization mechanism. Proc. Natl. Acad. Sci. U.S.A. 2017, 114, E2608. (PMID: 28289214) | |
 | Bos taurus (Bovine) (P02699) | 3.2-Angstrom In situ Mylar structure of bovine opsin at 100 K PDB File: | X-Ray Diffraction | 3.2 | 2017-12-13 | Broecker et al. High-throughput in situ X-ray screening of and data collection from protein crystals at room temperature and under cryogenic conditions. Nat Protoc. 2018, 13, 260. (PMID: 29300389) | |
 | Bos taurus (Bovine) (P02699) | Crystal structure of N2C/D282C stabilized opsin bound to RS01 PDB File: | X-Ray Diffraction | 2.36 | 2018-04-04 | Mattle et al. Ligand channel in pharmacologically stabilized rhodopsin. Proc. Natl. Acad. Sci. U.S.A. 2018, 115, 3640. (PMID: 29555765) | |
 | Bos taurus (Bovine) (P02699) | Crystal structure of N2C/D282C stabilized opsin bound to RS06 PDB File: | X-Ray Diffraction | 2.62 | 2018-04-04 | Mattle et al. Ligand channel in pharmacologically stabilized rhodopsin. Proc. Natl. Acad. Sci. U.S.A. 2018, 115, 3640. (PMID: 29555765) | |
 | Bos taurus (Bovine) (P02699) | Crystal structure of N2C/D282C stabilized opsin bound to RS08 PDB File: | X-Ray Diffraction | 2.87 | 2018-04-04 | Mattle et al. Ligand channel in pharmacologically stabilized rhodopsin. Proc. Natl. Acad. Sci. U.S.A. 2018, 115, 3640. (PMID: 29555765) | |
 | Bos taurus (Bovine) (P02699) | Crystal structure of N2C/D282C stabilized opsin bound to RS09 PDB File: | X-Ray Diffraction | 2.63 | 2018-04-04 | Mattle et al. Ligand channel in pharmacologically stabilized rhodopsin. Proc. Natl. Acad. Sci. U.S.A. 2018, 115, 3640. (PMID: 29555765) | |
 | Bos taurus (Bovine) (P02699) | Crystal structure of N2C/D282C stabilized opsin bound to RS11 PDB File: | X-Ray Diffraction | 2.7 | 2018-04-04 | Mattle et al. Ligand channel in pharmacologically stabilized rhodopsin. Proc. Natl. Acad. Sci. U.S.A. 2018, 115, 3640. (PMID: 29555765) | |
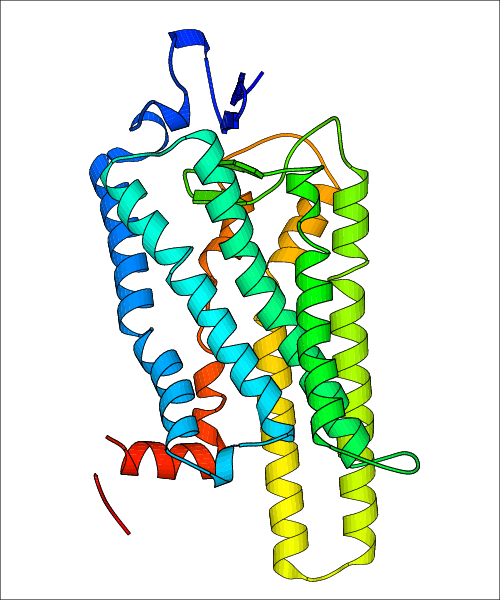 | Bos taurus (Bovine) (P02699) | Crystal structure of N2C/D282C stabilized opsin bound to RS13 PDB File: | X-Ray Diffraction | 3.03 | 2018-04-04 | Mattle et al. Ligand channel in pharmacologically stabilized rhodopsin. Proc. Natl. Acad. Sci. U.S.A. 2018, 115, 3640. (PMID: 29555765) | |
 | Bos taurus (Bovine) (P02699) | Crystal structure of N2C/D282C stabilized opsin bound to RS15 PDB File: | X-Ray Diffraction | 2.46 | 2018-04-04 | Mattle et al. Ligand channel in pharmacologically stabilized rhodopsin. Proc. Natl. Acad. Sci. U.S.A. 2018, 115, 3640. (PMID: 29555765) | |
 | Bos taurus (Bovine) (P02699) | Crystal structure of N2C/D282C stabilized opsin bound to RS16 PDB File: | X-Ray Diffraction | 2.49 | 2018-04-04 | Mattle et al. Ligand channel in pharmacologically stabilized rhodopsin. Proc. Natl. Acad. Sci. U.S.A. 2018, 115, 3640. (PMID: 29555765) | |
 | Bos taurus (Bovine) (P02699) | Crystal structure of the rhodopsin-mini-Go complex PDB File: | X-Ray Diffraction | 3.12 | 2018-10-03 | Tsai et al. Crystal structure of rhodopsin in complex with a mini-Gosheds light on the principles of G protein selectivity. Sci Adv. 2018, 4, eaat7052. (PMID: 30255144) | |
 | Bos taurus (Bovine) (P02699) | Cryo-EM structure of the native rhodopsin dimer from rod photoreceptor cells PDB File: | Electron Microscopy | 4.5 | 2019-08-21 | Zhao et al. Cryo-EM structure of the native rhodopsin dimer in nanodiscs. J.Biol.Chem. 2019. (PMID: 31399513) | |
 | Bos taurus (Bovine) (P02699) | Structure of the Rhodopsin-Transducin Complex PDB File: | Electron Microscopy | 3.9 | 2019-07-24 | Gao et al. Structures of the Rhodopsin-Transducin Complex: Insights into G-Protein Activation. Mol.Cell. 2019. (PMID: 31300275) | |
 | Bos taurus (Bovine) (P02699) | Structure of the Rhodopsin-Transducin-Nanobody Complex PDB File: | Electron Microscopy | 3.3 | 2019-07-24 | Gao et al. Structures of the Rhodopsin-Transducin Complex: Insights into G-Protein Activation. Mol.Cell. 2019. (PMID: 31300275) | |
 | Bos taurus (Bovine) (P02699) | Rhodopsin-Gi protein complex PDB File: | Electron Microscopy | 4.38 | 2019-07-10 | Tsai et al. Cryo-EM structure of the rhodopsin-G alpha i-beta gamma complex reveals binding of the rhodopsin C-terminal tail to the G beta subunit. Elife. 2019, 8, . (PMID: 31251171) | |
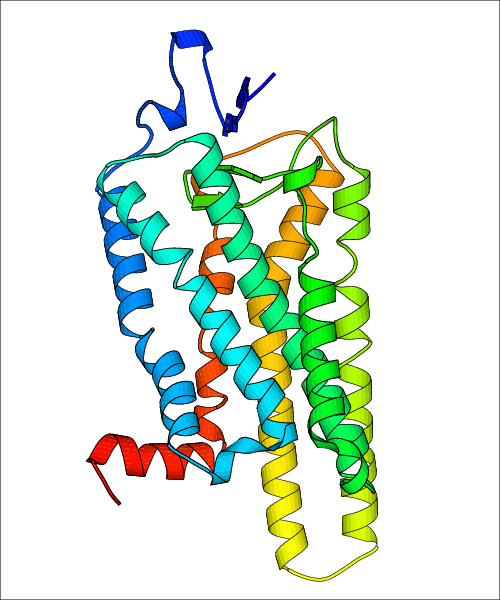 | Homo sapiens (Human) (P08100) | Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser PDB File: | X-Ray Diffraction | 3.3 | 2015-07-29 | Kang et al. Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser. Nature. 2015, 523, 561. (PMID: 26200343) | |
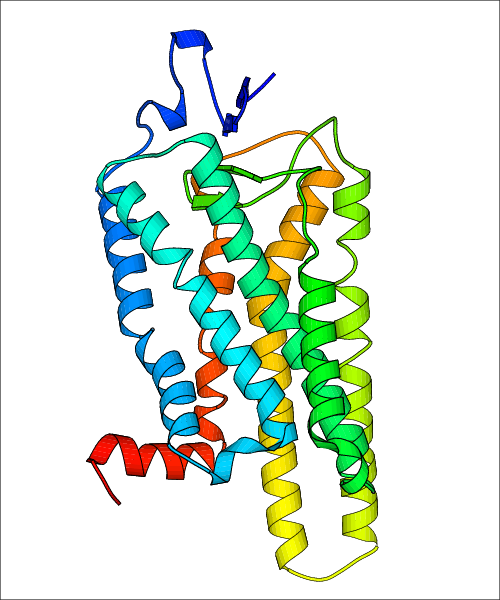 | Homo sapiens (Human) (P08100) | Crystal structure of rhodopsin bound to visual arrestin PDB File: | X-Ray Diffraction | 7.7 | 2016-03-23 | Zhou et al. X-ray laser diffraction for structure determination of the rhodopsin-arrestin complex. Sci Data. 2016, 3, 160021. (PMID: 27070998) | |
 | Homo sapiens (Human) (P08100) | Crystal structure of rhodopsin bound to visual arrestin determined by X-ray free electron laser PDB File: | X-Ray Diffraction | 3.01 | 2017-08-09 | Zhou et al. Identification of Phosphorylation Codes for Arrestin Recruitment by G Protein-Coupled Receptors. Cell. 2017, 170, 457. (PMID: 28753425) | |
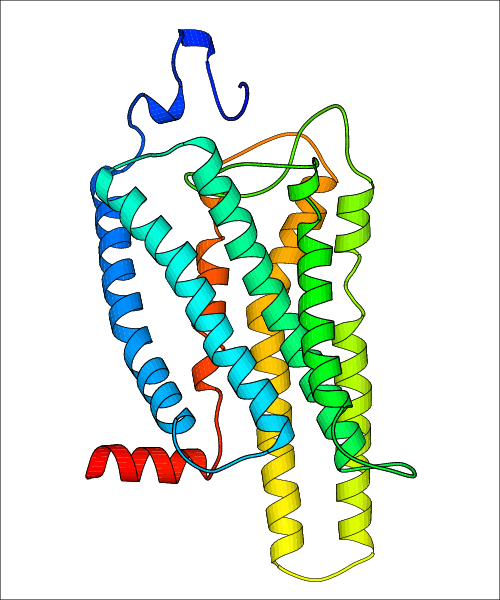 | Homo sapiens (Human) (P08100) | Rhodopsin-Gi complex PDB File: | Electron Microscopy | 4.5 | 2018-06-20 | Kang et al. Cryo-EM structure of human rhodopsin bound to an inhibitory G protein. Nature. 2018, 558, 553. (PMID: 29899450) | |
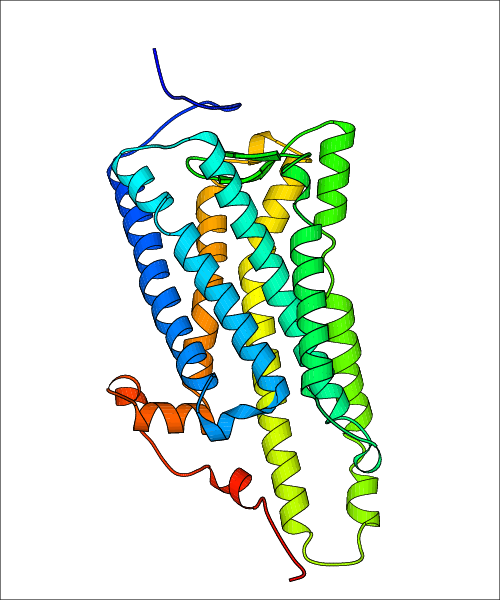 | Todarodes pacificus (Japanese flying squid) (P31356) | Crystal structure of squid rhodopsin PDB File: | X-Ray Diffraction | 2.5 | 2008-05-13 | Murakami et al. Crystal structure of squid rhodopsin. Nature. 2008, 453, 363. (PMID: 18480818) | |
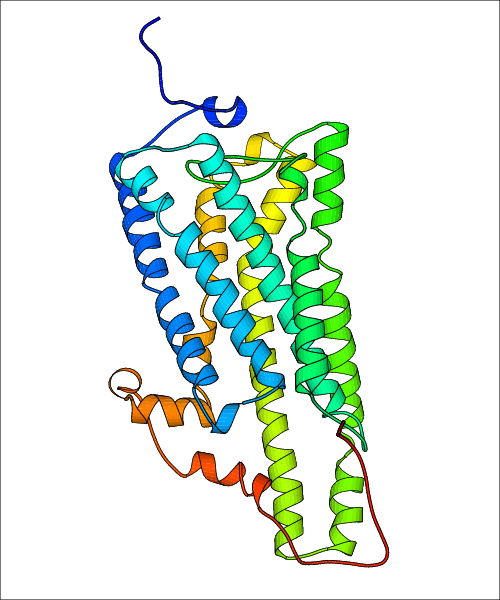 | Todarodes pacificus (Japanese flying squid) (P31356) | Crystal structure of squid rhodopsin PDB File: | X-Ray Diffraction | 3.7 | 2008-05-06 | Shimamura et al. Crystal structure of squid rhodopsin with intracellularly extended cytoplasmic region. J.Biol.Chem. 2008, 283, 17753. (PMID: 18463093) | |
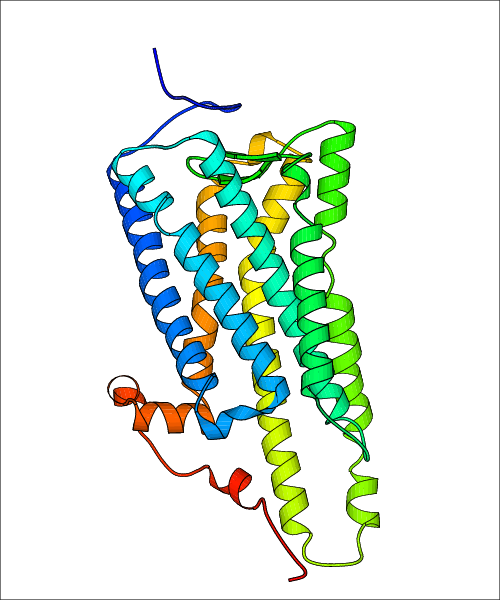 | Todarodes pacificus (Japanese flying squid) (P31356) | Crystal structure of the batho intermediate of squid rhodopsin PDB File: | X-Ray Diffraction | 2.8 | 2011-08-17 | Murakami et al. Crystallographic Analysis of the Primary Photochemical Reaction of Squid Rhodopsin. J.Mol.Biol. 2011, 413, 615. (PMID: 21906602) | |
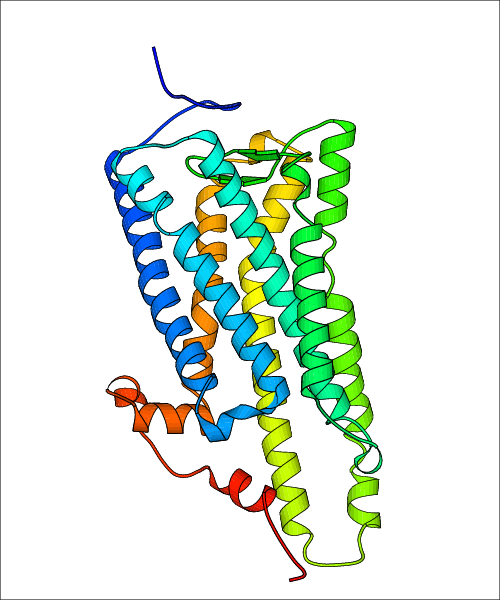 | Todarodes pacificus (Japanese flying squid) (P31356) | Crystal structure of squid isorhodopsin PDB File: | X-Ray Diffraction | 2.7 | 2011-08-17 | Murakami et al. Crystallographic Analysis of the Primary Photochemical Reaction of Squid Rhodopsin. J.Mol.Biol. 2011, 413, 615. (PMID: 21906602) | |
 | Todarodes pacificus (Japanese flying squid) (P31356) | Crystal structure of the lumi intermediate of squid rhodopsin PDB File: | X-Ray Diffraction | 2.8 | 2015-06-17 | Murakami et al. Crystallographic Study of the LUMI Intermediate of Squid Rhodopsin. Plos One. 2015, 10, e0126970. (PMID: 26024518) |
| PDB ID | Species (UniProt ID) | Description | Method | Resolution (Å) | Released | Reference | |
|---|---|---|---|---|---|---|---|
 | Homo sapiens (Human) (P21453) | Crystal Structure of a Lipid G protein-Coupled Receptor at 3.35A PDB File: | X-Ray Diffraction | 3.35 | 2012-02-15 | Hanson et al. Crystal structure of a lipid G protein-coupled receptor. Science. 2012, 335, 851. (PMID: 22344443) | |
 | Homo sapiens (Human) (P21453) | Crystal Structure of a Lipid G protein-Coupled Receptor at 2.80A PDB File: | X-Ray Diffraction | 2.8 | 2012-02-15 | Hanson et al. Crystal structure of a lipid G protein-coupled receptor. Science. 2012, 335, 851. (PMID: 22344443) |
| PDB ID | Species (UniProt ID) | Description | Method | Resolution (Å) | Released | Reference | |
|---|---|---|---|---|---|---|---|
 | Homo sapiens (Human) (P25103) | Solution conformation of substance P in water complexed with NK1R PDB File: | Solution NMR | N/A | 2010-11-03 | Gayen et al. NMR evidence of GM1-induced conformational change of Substance P using isotropic bicelles. Biochim.Biophys.Acta. 2010. (PMID: 20937248) | |
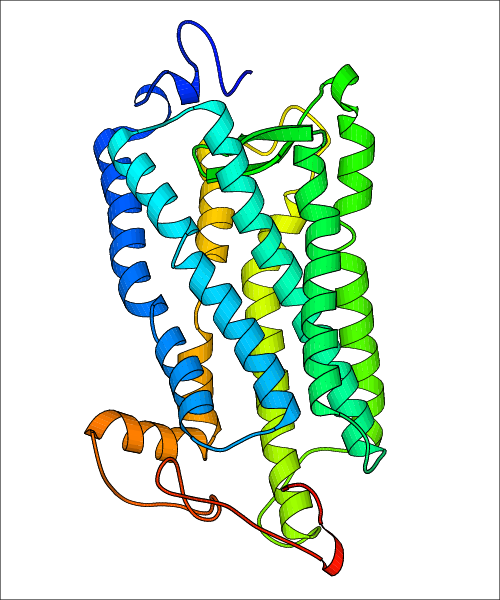 | Homo sapiens (Human) (P25103) | Substance P in DMPC/CHAPS isotropic q=0.25 bicelles as a ligand for NK1R PDB File: | Solution NMR | N/A | 2010-11-03 | Gayen et al. NMR evidence of GM1-induced conformational change of Substance P using isotropic bicelles. Biochim.Biophys.Acta. 2010. (PMID: 20937248) | |
 | Homo sapiens (Human) (P25103) | Substance P in isotropic q=0.25 DMPC/CHAPS/GM1 bicelles as a ligand for NK1R PDB File: | Solution NMR | N/A | 2010-11-03 | Gayen et al. NMR evidence of GM1-induced conformational change of Substance P using isotropic bicelles. Biochim.Biophys.Acta. 2010. (PMID: 20937248) | |
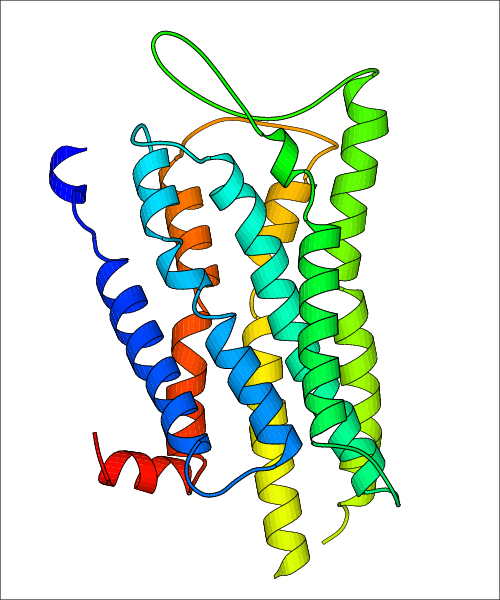 | Homo sapiens (Human) (P25103) | Crystal structure of the human NK1 tachykinin receptor PDB File: | X-Ray Diffraction | 3.4 | 2018-12-12 | Yin et al. Crystal structure of the human NK1tachykinin receptor. Proc. Natl. Acad. Sci. U.S.A. 2018, 115, 13264. (PMID: 30538204) | |
 | Homo sapiens (Human) (P25103) | Crystal structure of the Neurokinin 1 receptor in complex with the small molecule antagonist CP-99,994 PDB File: | X-Ray Diffraction | 3.27 | 2019-01-16 | Schoppe et al. Crystal structures of the human neurokinin 1 receptor in complex with clinically used antagonists. Nat Commun. 2019, 10, 17. (PMID: 30604743) | |
 | Homo sapiens (Human) (P25103) | Crystal structure of the Neurokinin 1 receptor in complex with the small molecule antagonist Aprepitant PDB File: | X-Ray Diffraction | 2.4 | 2019-01-16 | Schoppe et al. Crystal structures of the human neurokinin 1 receptor in complex with clinically used antagonists. Nat Commun. 2019, 10, 17. (PMID: 30604743) | |
 | Homo sapiens (Human) (P25103) | Crystal structure of the Neurokinin 1 receptor in complex with the small molecule antagonist Netupitant PDB File: | X-Ray Diffraction | 2.2 | 2019-01-16 | Schoppe et al. Crystal structures of the human neurokinin 1 receptor in complex with clinically used antagonists. Nat Commun. 2019, 10, 17. (PMID: 30604743) | |
 | Homo sapiens (Human) (P25103) | Crystal structure of the human NK1 substance P receptor PDB File: | X-Ray Diffraction | 2.7 | 2019-03-06 | Chen et al. Human substance P receptor binding mode of the antagonist drug aprepitant by NMR and crystallography. Nat Commun. 2019, 10, 638. (PMID: 30733446) | |
 | Homo sapiens (Human) (P25103) | Crystal structure of the human NK1 substance P receptor PDB File: | X-Ray Diffraction | 3.2 | 2019-03-06 | Chen et al. Human substance P receptor binding mode of the antagonist drug aprepitant by NMR and crystallography. Nat Commun. 2019, 10, 638. (PMID: 30733446) |
| PDB ID | Species (UniProt ID) | Description | Method | Resolution (Å) | Released | Reference | |
|---|---|---|---|---|---|---|---|
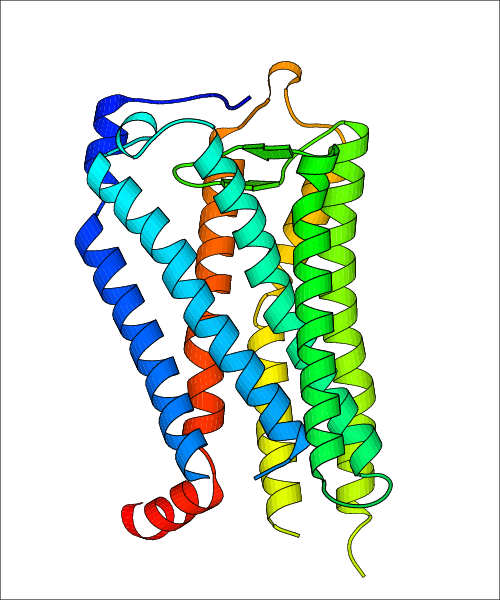 | Homo sapiens (Human) (P21731) | Crystal structure of the human thromboxane A2 receptor bound to ramatroban PDB File: | X-Ray Diffraction | 2.5 | 2018-12-19 | Fan et al. Structural basis for ligand recognition of the human thromboxane A2receptor. Nat. Chem. Biol. 2019, 15, 27. (PMID: 30510189) | |
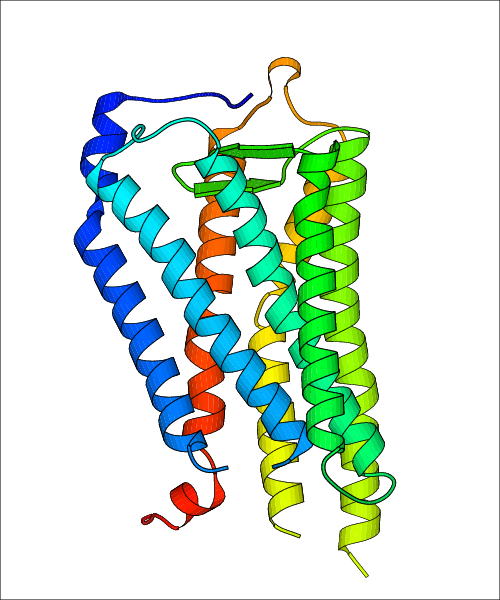 | Homo sapiens (Human) (P21731) | Crystal structure of the human thromboxane A2 receptor bound to daltroban PDB File: | X-Ray Diffraction | 3.0 | 2018-12-19 | Fan et al. Structural basis for ligand recognition of the human thromboxane A2receptor. Nat. Chem. Biol. 2019, 15, 27. (PMID: 30510189) |
| PDB ID | Species (UniProt ID) | Description | Method | Resolution (Å) | Released | Reference | |
|---|---|---|---|---|---|---|---|
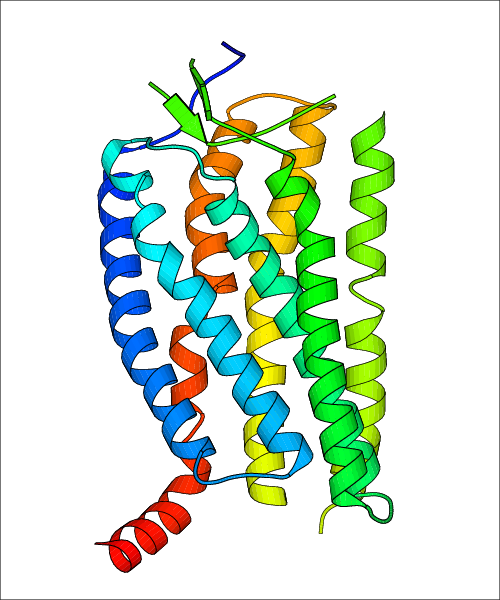 | Homo sapiens (Human) (P30556) | XFEL structure of human Angiotensin Receptor PDB File: | X-Ray Diffraction | 2.9 | 2015-04-22 | Zhang et al. Structure of the Angiotensin receptor revealed by serial femtosecond crystallography. Cell. 2015, 161, 833. (PMID: 25913193) | |
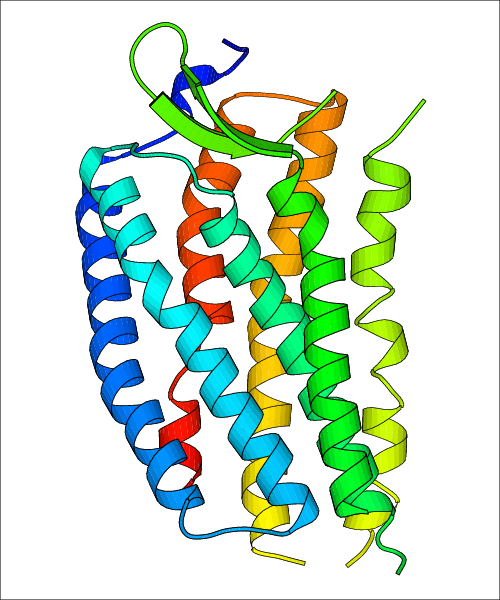 | Homo sapiens (Human) (P30556) | Crystal Structure of Human Angiotensin Receptor in Complex with Inverse Agonist Olmesartan at 2.8A resolution. PDB File: | X-Ray Diffraction | 2.8 | 2015-10-07 | Zhang et al. Structural Basis for Ligand Recognition and Functional Selectivity at Angiotensin Receptor. J.Biol.Chem. 2015, 290, 29127. (PMID: 26420482) | |
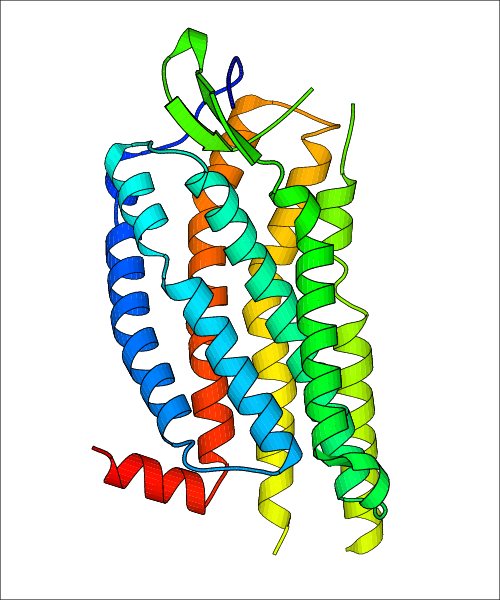 | Homo sapiens (Human) (P30556) | Structure of nanobody-stabilized angiotensin II type 1 receptor bound to S1I8 PDB File: | X-Ray Diffraction | 2.9 | 2019-01-30 | Wingler et al. Distinctive Activation Mechanism for Angiotensin Receptor Revealed by a Synthetic Nanobody. Cell. 2019. (PMID: 30639100) |
| PDB ID | Species (UniProt ID) | Description | Method | Resolution (Å) | Released | Reference | |
|---|---|---|---|---|---|---|---|
 | Homo sapiens (Human) (P50052) | XFEL structure of human angiotensin II type 2 receptor (Monoclinic form) in complex with compound 1 (N-benzyl-N-(2-ethyl-4-oxo-3-{[2'-(2H-tetrazol-5-yl)[1,1'-biphenyl]-4-yl]) PDB File: | X-Ray Diffraction | 2.8 | 2017-04-05 | Zhang et al. Structural basis for selectivity and diversity in angiotensin II receptors. Nature. 2017, 544, 327. (PMID: 28379944) | |
 | Homo sapiens (Human) (P50052) | XFEL structure of human angiotensin II type 2 receptor (Orthorhombic form) in complex with compound 1 (N-benzyl-N-(2-ethyl-4-oxo-3-{[2'-(2H-tetrazol-5-yl)[1,1'-biphenyl]-4-yl] methyl}-3,4-dihydroquinazolin-6-yl)thiophene-2-carboxamide) PDB File: | X-Ray Diffraction | 2.8 | 2017-04-05 | Zhang et al. Structural basis for selectivity and diversity in angiotensin II receptors. Nature. 2017, 544, 327. (PMID: 28379944) | |
 | Homo sapiens (Human) (P50052) | Synchrotron structure of human angiotensin II type 2 receptor in complex with compound 2 (N-[(furan-2-yl)methyl]-N-(4-oxo-2-propyl-3-{[2'-(2H-tetrazol-5-yl)[1,1'- biphenyl]-4-yl]methyl}-3,4-dihydroquinazolin-6-yl)benzamide) PDB File: | X-Ray Diffraction | 2.9 | 2017-04-05 | Zhang et al. Structural basis for selectivity and diversity in angiotensin II receptors. Nature. 2017, 544, 327. (PMID: 28379944) | |
 | Homo sapiens (Human) (P50052) | Complex structure of angiotensin II type 2 receptor with Fab PDB File: | X-Ray Diffraction | 3.2 | 2018-07-11 | Asada et al. Crystal structure of the human angiotensin II type 2 receptor bound to an angiotensin II analog. Nat. Struct. Mol. Biol. 2018. (PMID: 29967536) |
| PDB ID | Species (UniProt ID) | Description | Method | Resolution (Å) | Released | Reference | |
|---|---|---|---|---|---|---|---|
 | Homo sapiens (Human) (Q16602) | Cryo-EM structure of the active, Gs-protein complexed, human CGRP receptor PDB File: | Electron Microscopy | 3.3 | 2018-09-19 | Liang et al. Cryo-EM structure of the active, Gs-protein complexed, human CGRP receptor. Nature. 2018, 561, 492. (PMID: 30209400) |
| PDB ID | Species (UniProt ID) | Description | Method | Resolution (Å) | Released | Reference | |
|---|---|---|---|---|---|---|---|
 | Homo sapiens (Human) (P30988) | Volta phase plate cryo-electron microscopy structure of a calcitonin receptor-heterotrimeric Gs protein complex PDB File: | Electron Microscopy | 4.1 | 2017-05-03 | Liang et al. Phase-plate cryo-EM structure of a class B GPCR-G-protein complex. Nature. 2017, 546, 118. (PMID: 28437792) | |
 | Homo sapiens (Human) (P30988) | A high-resolution cryo-electron microscopy structure of a calcitonin receptor-heterotrimeric Gs protein complex PDB File: | Electron Microscopy | 3.34 | 2019-01-23 | dal Maso et al. The Molecular Control of Calcitonin Receptor Signaling. Acs Pharmacol Transl Sci. 2019, , 31. (PMID: N/A) |
| PDB ID | Species (UniProt ID) | Description | Method | Resolution (Å) | Released | Reference | |
|---|---|---|---|---|---|---|---|
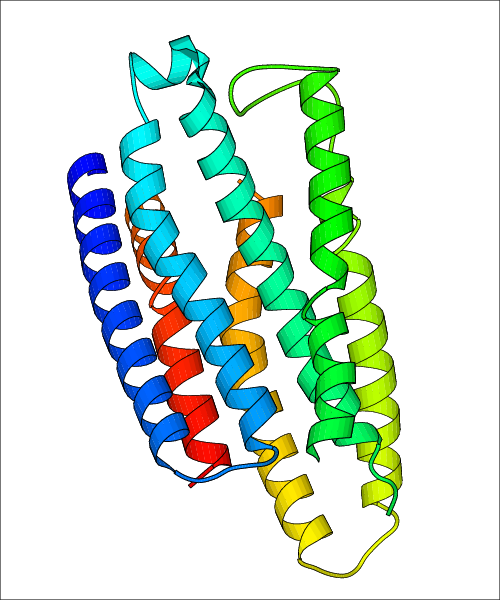 | Homo sapiens (Human) (P34998) | Crystal structure of human corticotropin-releasing factor receptor 1 (CRF1R) in complex with the antagonist CP-376395 PDB File: | X-Ray Diffraction | 2.98 | 2013-07-17 | Hollenstein et al. Structure of class B GPCR corticotropin-releasing factor receptor 1. Nature. 2013, 499, 438. (PMID: 23863939) | |
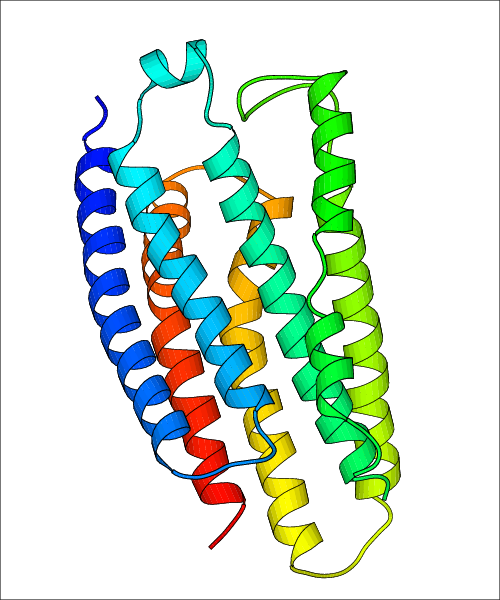 | Homo sapiens (Human) (P34998) | Crystal structure of human corticotropin-releasing factor receptor 1 (CRF1R) in complex with the antagonist CP-376395 in a hexagonal setting with translational non-crystallographic symmetry PDB File: | X-Ray Diffraction | 3.18 | 2016-06-29 | Dore et al. Decoding Corticotropin-Releasing Factor Receptor Type 1 Crystal Structures. Curr Mol Pharmacol. 2017, 10, 334. (PMID: 28183242) |
| PDB ID | Species (UniProt ID) | Description | Method | Resolution (Å) | Released | Reference | |
|---|---|---|---|---|---|---|---|
 | Homo sapiens (Human) (P47871) | Structure of the class B human glucagon G protein coupled receptor PDB File: | X-Ray Diffraction | 3.3 | 2013-07-24 | Siu et al. Structure of the human glucagon class B G-protein-coupled receptor. Nature. 2013, 499, 444. (PMID: 23863937) | |
 | Homo sapiens (Human) (P47871) | Crystal structure of the human glucagon receptor (GCGR) in complex with the antagonist MK-0893 PDB File: | X-Ray Diffraction | 2.5 | 2016-04-20 | Jazayeri et al. Extra-helical binding site of a glucagon receptor antagonist. Nature. 2016, 533, 274. (PMID: 27111510) | |
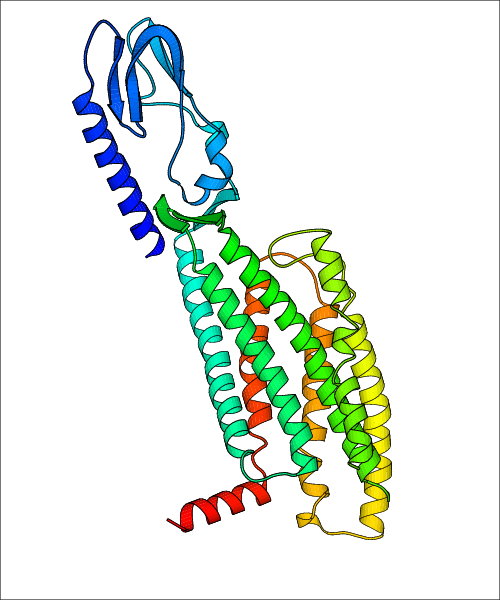 | Homo sapiens (Human) (P47871) | Structure of the Full-length glucagon class B G protein-coupled receptor PDB File: | X-Ray Diffraction | 3.0 | 2017-05-24 | Zhang et al. Structure of the full-length glucagon class B G-protein-coupled receptor. Nature. 2017, 546, 259. (PMID: 28514451) | |
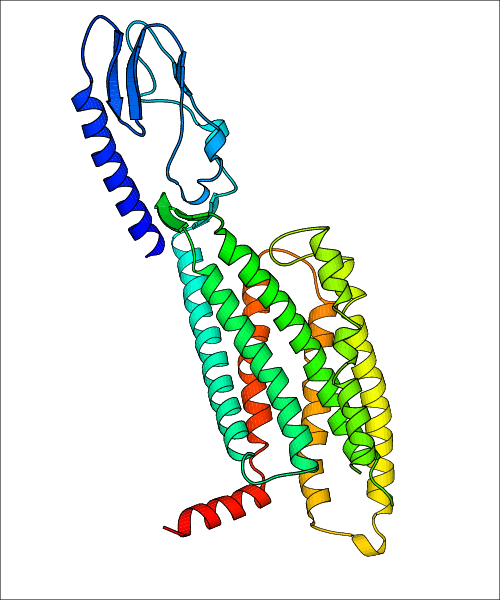 | Homo sapiens (Human) (P47871) | Structure of the Full-length glucagon class B G protein-coupled receptor PDB File: | X-Ray Diffraction | 3.19 | 2017-05-24 | Zhang et al. Structure of the full-length glucagon class B G-protein-coupled receptor. Nature. 2017, 546, 259. (PMID: 28514451) | |
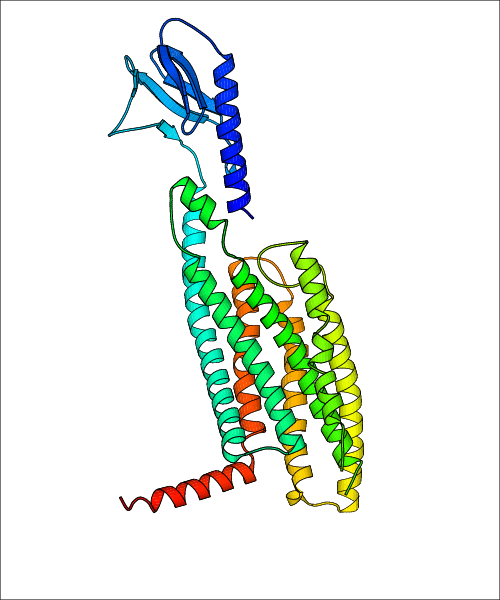 | Homo sapiens (Human) (P47871) | Structure of the glucagon receptor in complex with a glucagon analogue PDB File: | X-Ray Diffraction | 3.0 | 2018-01-17 | Zhang et al. Structure of the glucagon receptor in complex with a glucagon analogue. Nature. 2018, 553, 106. (PMID: 29300013) |
| PDB ID | Species (UniProt ID) | Description | Method | Resolution (Å) | Released | Reference | |
|---|---|---|---|---|---|---|---|
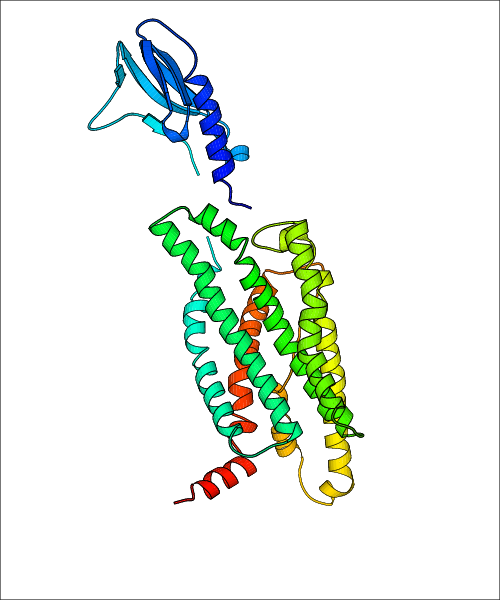 | Oryctolagus cuniculus (Rabbit) (G1SGD4) | Cryo-EM structure of the activated Glucagon-like peptide-1 receptor in complex with G protein PDB File: | Electron Microscopy | 4.1 | 2017-05-24 | Zhang et al. Cryo-EM structure of the activated GLP-1 receptor in complex with a G protein. Nature. 2017, 546, 248. (PMID: 28538729) | |
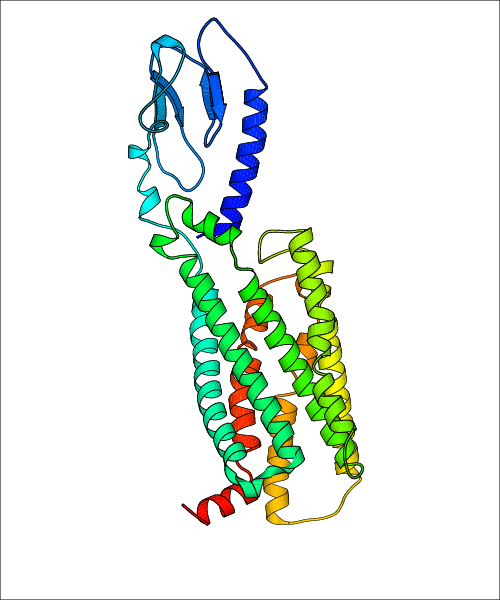 | Homo sapiens (Human) (P43220) | Crystal structure of thermostabilised full-length GLP-1R in complex with a truncated peptide agonist at 3.7 A resolution PDB File: | X-Ray Diffraction | 3.7 | 2017-06-14 | Jazayeri et al. Crystal structure of the GLP-1 receptor bound to a peptide agonist. Nature. 2017, 546, 254. (PMID: 28562585) | |
 | Homo sapiens (Human) (P43220) | Structure of the human GLP-1 receptor complex with PF-06372222 PDB File: | X-Ray Diffraction | 2.7 | 2017-05-24 | Song et al. Human GLP-1 receptor transmembrane domain structure in complex with allosteric modulators. Nature. 2017, 546, 312. (PMID: 28514449) | |
 | Homo sapiens (Human) (P43220) | Structure of the human GLP-1 receptor complex with NNC0640 PDB File: | X-Ray Diffraction | 3.0 | 2017-05-17 | Song et al. Human GLP-1 receptor transmembrane domain structure in complex with allosteric modulators. Nature. 2017, 546, 312. (PMID: 28514449) | |
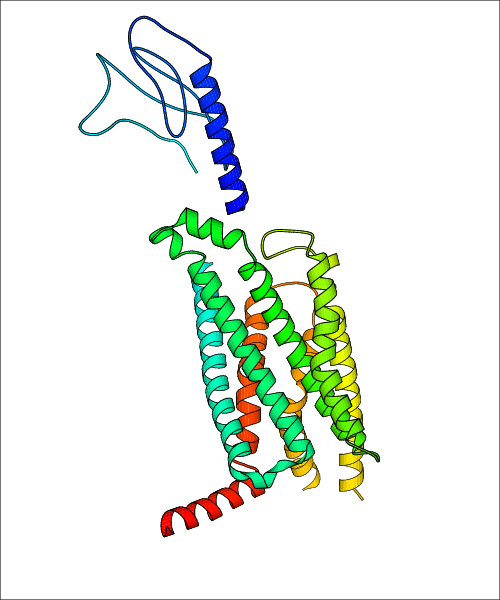 | Homo sapiens (Human) (P43220) | 3.3 angstrom phase-plate cryo-EM structure of a biased agonist-bound human GLP-1 receptor-Gs complex PDB File: | Electron Microscopy | 3.3 | 2018-02-21 | Liang et al. Phase-plate cryo-EM structure of a biased agonist-bound human GLP-1 receptor-Gs complex. Nature. 2018, 555, 121. (PMID: 29466332) | |
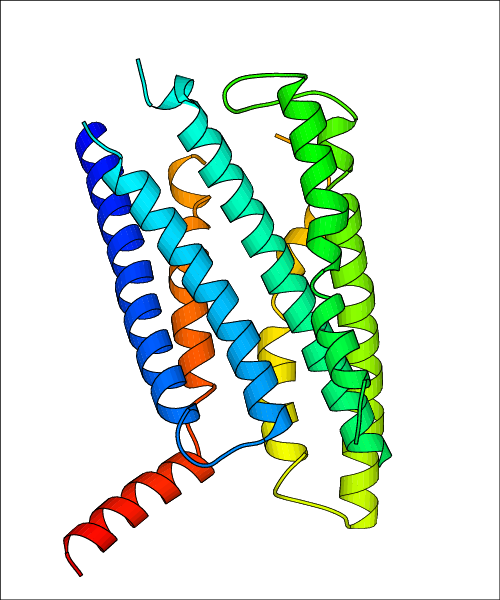 | Homo sapiens (Human) (P43220) | Structure of thermal-stabilised(M9) human GLP-1 receptor transmembrane domain PDB File: | X-Ray Diffraction | 2.8 | 2019-11-13 | Xu et al. Mutagenesis facilitated crystallization of GLP-1R. Iucrj. 2019, 6, 996. (PMID: 31709055) | |
 | Homo sapiens (Human) (P43220) | Structure of thermal-stabilised(M8) human GLP-1 receptor transmembrane domain PDB File: | X-Ray Diffraction | 2.8 | 2019-11-13 | Xu et al. Mutagenesis facilitated crystallization of GLP-1R. Iucrj. 2019, 6, 996. (PMID: 31709055) | |
 | Homo sapiens (Human) (P43220) | Structure of thermal-stabilised(M6) human GLP-1 receptor transmembrane domain PDB File: | X-Ray Diffraction | 3.1 | 2019-11-13 | Xu et al. Mutagenesis facilitated crystallization of GLP-1R. Iucrj. 2019, 6, 996. (PMID: 31709055) |
| PDB ID | Species (UniProt ID) | Description | Method | Resolution (Å) | Released | Reference | |
|---|---|---|---|---|---|---|---|
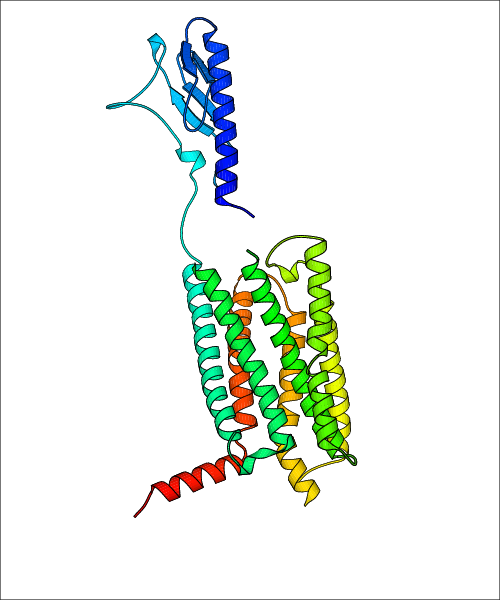 | Homo sapiens (Human) (Q03431) | High resolution crystal structure of parathyroid hormone 1 receptor in complex with a peptide agonist. PDB File: | X-Ray Diffraction | 2.5 | 2018-11-21 | Ehrenmann et al. High-resolution crystal structure of parathyroid hormone 1 receptor in complex with a peptide agonist. Nat. Struct. Mol. Biol. 2018, 25, 1086. (PMID: 30455434) | |
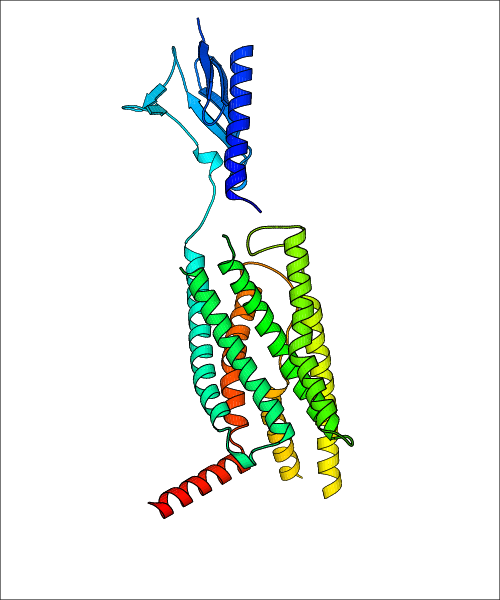 | Homo sapiens (Human) (Q03431) | Cryo-EM structure of parathyroid hormone receptor type 1 in complex with a long-acting parathyroid hormone analog and G protein PDB File: | Electron Microscopy | 3.0 | 2019-04-17 | Zhao et al. Structure and dynamics of the active human parathyroid hormone receptor-1. Science. 2019, 364, 148. (PMID: 30975883) | |
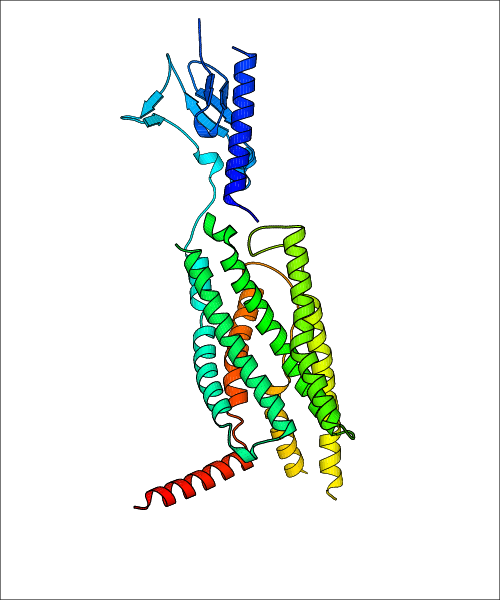 | Homo sapiens (Human) (Q03431) | Cryo-EM structure of parathyroid hormone receptor type 1 in complex with a long-acting parathyroid hormone analog and G protein PDB File: | Electron Microscopy | 3.5 | 2019-04-17 | Zhao et al. Structure and dynamics of the active human parathyroid hormone receptor-1. Science. 2019, 364, 148. (PMID: 30975883) | |
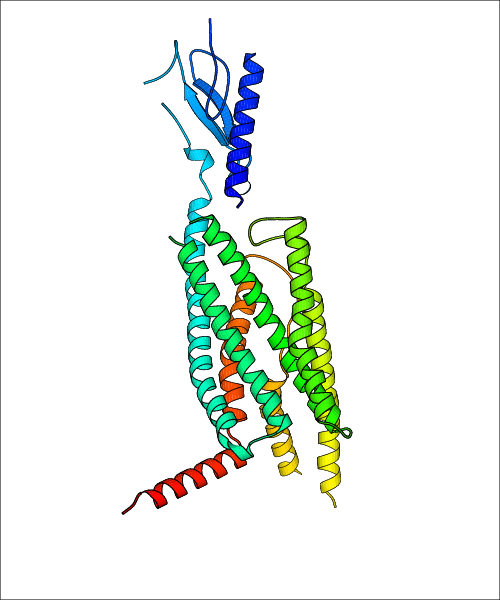 | Homo sapiens (Human) (Q03431) | Cryo-EM structure of parathyroid hormone receptor type 1 in complex with a long-acting parathyroid hormone analog and G protein PDB File: | Electron Microscopy | 4.0 | 2019-04-17 | Zhao et al. Structure and dynamics of the active human parathyroid hormone receptor-1. Science. 2019, 364, 148. (PMID: 30975883) |
| PDB ID | Species (UniProt ID) | Description | Method | Resolution (Å) | Released | Reference | |
|---|---|---|---|---|---|---|---|
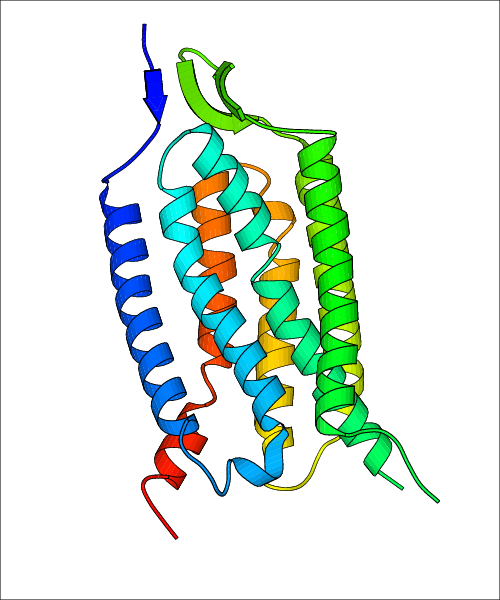 | Homo sapiens (Human) (Q13255) | Human class C G protein-coupled metabotropic glutamate receptor 1 in complex with a negative allosteric modulator PDB File: | X-Ray Diffraction | 2.8 | 2014-03-19 | Wu et al. Structure of a class C GPCR metabotropic glutamate receptor 1 bound to an allosteric modulator. Science. 2014, 344, 58. (PMID: 24603153) |
| PDB ID | Species (UniProt ID) | Description | Method | Resolution (Å) | Released | Reference | |
|---|---|---|---|---|---|---|---|
 | Homo sapiens (Human) (P41594) | Structure of the human class C GPCR metabotropic glutamate receptor 5 transmembrane domain in complex with the negative allosteric modulator mavoglurant PDB File: | X-Ray Diffraction | 2.6 | 2014-07-02 | Dore et al. Structure of class C GPCR metabotropic glutamate receptor 5 transmembrane domain. Nature. 2014, 511, 557. (PMID: 25042998) | |
 | Homo sapiens (Human) (P41594) | Structure of the human class C GPCR metabotropic glutamate receptor 5 transmembrane domain in complex with the negative allosteric modulator 3-chloro-4-fluoro-5-[6-(1H-pyrazol-1-yl)pyrimidin-4-yl]benzonitrile PDB File: | X-Ray Diffraction | 3.1 | 2015-08-12 | Christopher et al. Fragment and Structure-Based Drug Discovery for a Class C GPCR: Discovery of the mGlu5 Negative Allosteric Modulator HTL14242 (3-Chloro-5-[6-(5-fluoropyridin-2-yl)pyrimidin-4-yl]benzonitrile). J.Med.Chem. 2015, 58, 6653. (PMID: 26225459) | |
 | Homo sapiens (Human) (P41594) | Structure of the human class C GPCR metabotropic glutamate receptor 5 transmembrane domain in complex with the negative allosteric modulator 3-chloro-5-[6-(5-fluoropyridin-2-yl)pyrimidin-4-yl]benzonitrile - (HTL14242) PDB File: | X-Ray Diffraction | 2.6 | 2015-08-12 | Christopher et al. Fragment and Structure-Based Drug Discovery for a Class C GPCR: Discovery of the mGlu5 Negative Allosteric Modulator HTL14242 (3-Chloro-5-[6-(5-fluoropyridin-2-yl)pyrimidin-4-yl]benzonitrile). J.Med.Chem. 2015, 58, 6653. (PMID: 26225459) | |
 | Homo sapiens (Human) (P41594) | Crystal Structure of mGluR5 in complex with Fenobam at 2.65 A PDB File: | X-Ray Diffraction | 2.65 | 2018-03-07 | Christopher et al. Structure-Based Optimization Strategies for G Protein-Coupled Receptor (GPCR) Allosteric Modulators: A Case Study from Analyses of New Metabotropic Glutamate Receptor 5 (mGlu5) X-ray Structures. J. Med. Chem. 2018. (PMID: 29455526) | |
 | Homo sapiens (Human) (P41594) | Crystal Structure of mGluR5 in complex with MMPEP at 2.2 A PDB File: | X-Ray Diffraction | 2.2 | 2018-03-07 | Christopher et al. Structure-Based Optimization Strategies for G Protein-Coupled Receptor (GPCR) Allosteric Modulators: A Case Study from Analyses of New Metabotropic Glutamate Receptor 5 (mGlu5) X-ray Structures. J. Med. Chem. 2018. (PMID: 29455526) | |
 | Homo sapiens (Human) (P41594) | Metabotropic Glutamate Receptor 5 Apo Form Ligand Binding Domain PDB File: | X-Ray Diffraction | 4.0 | 2019-01-23 | Koehl et al. Structural insights into the activation of metabotropic glutamate receptors. Nature. 2019. (PMID: 30675062) | |
 | Homo sapiens (Human) (P41594) | Metabotropic Glutamate Receptor 5 bound to L-quisqualate and Nb43 PDB File: | Electron Microscopy | 4.0 | 2019-01-23 | Koehl et al. Structural insights into the activation of metabotropic glutamate receptors. Nature. 2019. (PMID: 30675062) | |
 | Homo sapiens (Human) (P41594) | Metabotropic Glutamate Receptor 5 Apo Form PDB File: | Electron Microscopy | 4.0 | 2019-01-23 | Koehl et al. Structural insights into the activation of metabotropic glutamate receptors. Nature. 2019. (PMID: 30675062) |
| PDB ID | Species (UniProt ID) | Description | Method | Resolution (Å) | Released | Reference | |
|---|---|---|---|---|---|---|---|
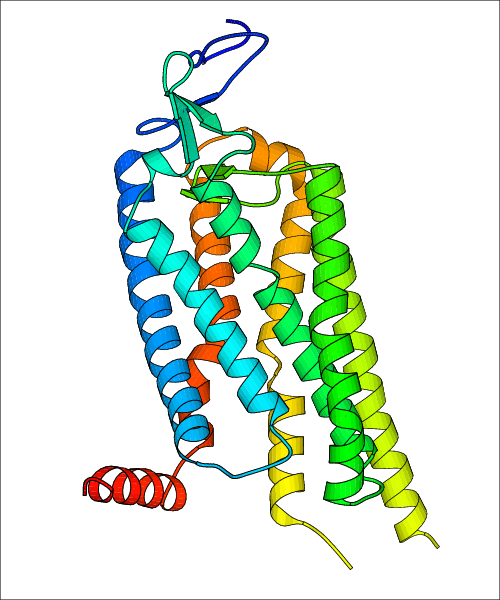 | Homo sapiens (Human) (Q9ULV1) | Crystal structure of human apo-Frizzled4 receptor PDB File: | X-Ray Diffraction | 2.4 | 2018-08-22 | Yang et al. Crystal structure of the Frizzled 4 receptor in a ligand-free state. Nature. 2018. (PMID: 30135577) |
| PDB ID | Species (UniProt ID) | Description | Method | Resolution (Å) | Released | Reference | |
|---|---|---|---|---|---|---|---|
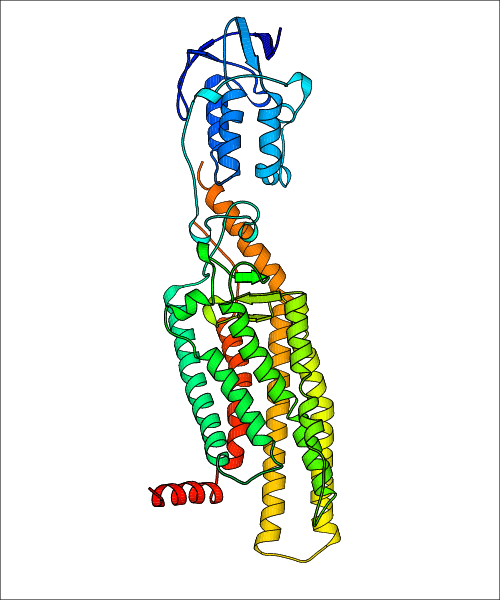 | Mus musculus (Mouse) (P56726) | Crystal structure of active Smoothened bound to SAG21k, cholesterol, and NbSmo8 PDB File: | X-Ray Diffraction | 2.8 | 2019-07-03 | Deshpande et al. Smoothened stimulation by membrane sterols drives Hedgehog pathway activity. Nature. 2019. (PMID: 31263273) | |
 | Homo sapiens (Human) (Q99835) | Structure of the human smoothened 7TM receptor in complex with an antitumor agent PDB File: | X-Ray Diffraction | 2.45 | 2013-04-24 | Wang et al. Structure of the human smoothened receptor bound to an antitumour agent. Nature. 2013, 497, 338. (PMID: 23636324) | |
 | Homo sapiens (Human) (Q99835) | Structure of the human smoothened receptor in complex with SANT-1. PDB File: | X-Ray Diffraction | 2.8 | 2014-01-22 | Wang et al. Structural basis for Smoothened receptor modulation and chemoresistance to anticancer drugs. Nat Commun. 2014, 5, 4355. (PMID: 25008467) | |
 | Homo sapiens (Human) (Q99835) | Human Smoothened Receptor structure in complex with cyclopamine PDB File: | X-Ray Diffraction | 3.2 | 2014-03-05 | Weierstall et al. Lipidic cubic phase injector facilitates membrane protein serial femtosecond crystallography. Nat Commun. 2014, 5, 3309. (PMID: 24525480) | |
 | Homo sapiens (Human) (Q99835) | Structure of the human smoothened receptor in complex with ANTA XV PDB File: | X-Ray Diffraction | 2.61 | 2014-07-23 | Wang et al. Structural basis for Smoothened receptor modulation and chemoresistance to anticancer drugs. Nat Commun. 2014, 5, 4355. (PMID: 25008467) | |
 | Homo sapiens (Human) (Q99835) | Structure of the human smoothened receptor in complex with SAG1.5 PDB File: | X-Ray Diffraction | 2.6 | 2014-07-23 | Wang et al. Structural basis for Smoothened receptor modulation and chemoresistance to anticancer drugs. Nat Commun. 2014, 5, 4355. (PMID: 25008467) | |
 | Homo sapiens (Human) (Q99835) | Structure of human Smoothened in complex with cholesterol PDB File: | X-Ray Diffraction | 3.2 | 2016-07-20 | Byrne et al. Structural basis of Smoothened regulation by its extracellular domains. Nature. 2016, 535, 517. (PMID: 27437577) | |
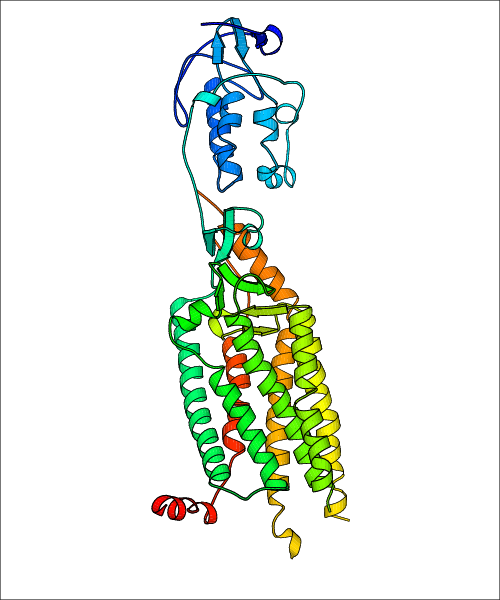 | Homo sapiens (Human) (Q99835) | Structure of human Smoothened in complex with Vismodegib PDB File: | X-Ray Diffraction | 3.3 | 2016-07-20 | Byrne et al. Structural basis of Smoothened regulation by its extracellular domains. Nature. 2016, 535, 517. (PMID: 27437577) | |
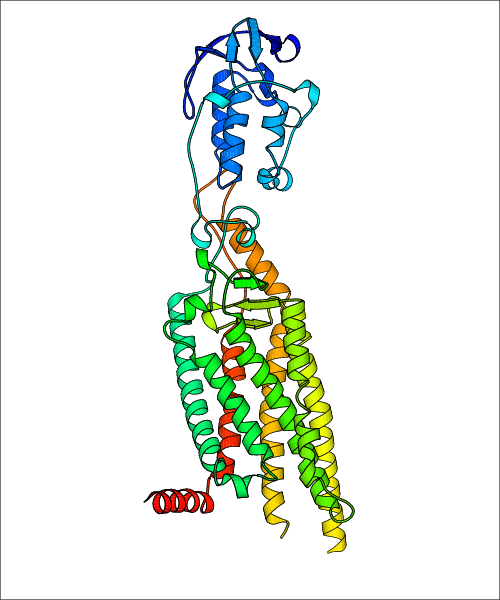 | Homo sapiens (Human) (Q99835) | 2.9A XFEL structure of the multi-domain human smoothened receptor (with E194M mutation) in complex with TC114 PDB File: | X-Ray Diffraction | 2.9 | 2017-05-24 | Zhang et al. Crystal structure of a multi-domain human smoothened receptor in complex with a super stabilizing ligand. Nat Commun. 2017, 8, 15383. (PMID: 28513578) | |
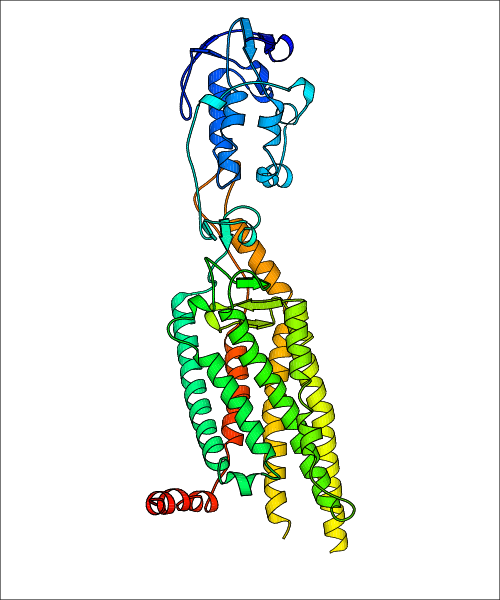 | Homo sapiens (Human) (Q99835) | 3.0A SYN structure of the multi-domain human smoothened receptor in complex with TC114 PDB File: | X-Ray Diffraction | 3.0 | 2017-05-24 | Zhang et al. Crystal structure of a multi-domain human smoothened receptor in complex with a super stabilizing ligand. Nat Commun. 2017, 8, 15383. (PMID: 28513578) | |
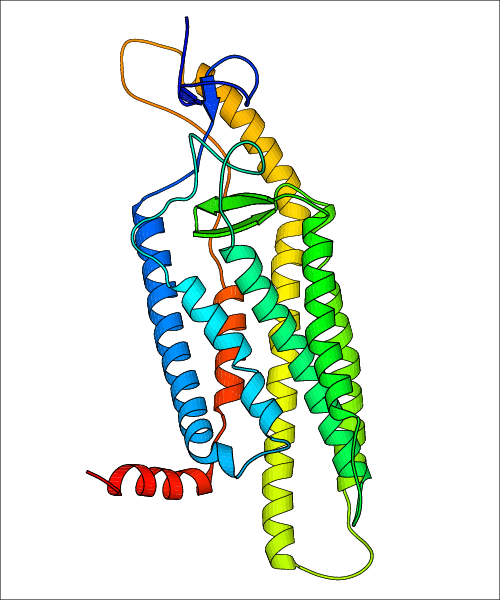 | Homo sapiens (Human) (Q99835) | Structure of human Smoothened-Gi complex PDB File: | Electron Microscopy | 3.84 | 2019-06-12 | Qi et al. Cryo-EM structure of oxysterol-bound human Smoothened coupled to a heterotrimeric Gi. Nature. 2019. (PMID: 31168089) |
yangzhanglab![]() umich.edu
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
umich.edu
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218


 1.) 5-hydroxytryptamine receptor 1B (n = 4)
1.) 5-hydroxytryptamine receptor 1B (n = 4)