

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 400 420 440 460 480 | |
| | | | | | | | | | | | | | | | | | | | | | | | | | |
| MRWLWPLAVSLAVILAVGLSRVSGGAPLHLGRHRAETQEQQSRSKRGTEDEEAKGVQQYVPEEWAEYPRPIHPAGLQPTKPLVATSPNPGKDGGTPDSGQELRGNLTGAPGQRLQIQNPLYPVTESSYSAYAIMLLALVVFAVGIVGNLSVMCIVWHSYYLKSAWNSILASLALWDFLVLFFCLPIVIFNEITKQRLLGDVSCRAVPFMEVSSLGVTTFSLCALGIDRFHVATSTLPKVRPIERCQSILAKLAVIWVGSMTLAVPELLLWQLAQEPAPTMGTLDSCIMKPSASLPESLYSLVMTYQNARMWWYFGCYFCLPILFTVTCQLVTWRVRGPPGRKSECRASKHEQCESQLNSTVVGLTVVYAFCTLPENVCNIVVAYLSTELTRQTLDLLGLINQFSTFFKGAITPVLLLCICRPLGQAFLDCCCCCCCEECGGASEASAANGSDNKLKTEVSSSIYFHKPRESPPLLPLGTPC | |
| CCCCCHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHCCHHHHSSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCSSCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHSSSSSSSSCCCCCCSSSSSSSCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC | |
| 9763337776778886133579998665544456667675555776333544467677876201478888798877889863216899999999998777777877899887765567777766531899999999999999984509660755684587776107988757999999999988999999994986463268887999999999999999999998716125211522535521898999999999999999999998189898514788714799876797543004135567999999999999899999999999999997827888743045889999989999999999999999999999999999984742335699999999999999999999999996599999999999503157877887877677789874105778888888857788878999999 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 400 420 440 460 480 | |
| | | | | | | | | | | | | | | | | | | | | | | | | | |
| MRWLWPLAVSLAVILAVGLSRVSGGAPLHLGRHRAETQEQQSRSKRGTEDEEAKGVQQYVPEEWAEYPRPIHPAGLQPTKPLVATSPNPGKDGGTPDSGQELRGNLTGAPGQRLQIQNPLYPVTESSYSAYAIMLLALVVFAVGIVGNLSVMCIVWHSYYLKSAWNSILASLALWDFLVLFFCLPIVIFNEITKQRLLGDVSCRAVPFMEVSSLGVTTFSLCALGIDRFHVATSTLPKVRPIERCQSILAKLAVIWVGSMTLAVPELLLWQLAQEPAPTMGTLDSCIMKPSASLPESLYSLVMTYQNARMWWYFGCYFCLPILFTVTCQLVTWRVRGPPGRKSECRASKHEQCESQLNSTVVGLTVVYAFCTLPENVCNIVVAYLSTELTRQTLDLLGLINQFSTFFKGAITPVLLLCICRPLGQAFLDCCCCCCCEECGGASEASAANGSDNKLKTEVSSSIYFHKPRESPPLLPLGTPC | |
| 4310000001002213222233453351524443444544544354246454344334223631353231241442433322222122333332223323313111122324334243212323322000000000010002001300100000001123021000000000000000000000100000000320001200300000000000000000000000000000000033333232220000000000000000000000002023242354310000001022622310000000123020000000000200000000000000113334544543445134211000000000000000010000000000000233123100000000000000200000000000004300400130000012344664444333434433343434434434334433343436437 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 400 420 440 460 480 | | | | | | | | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHCCHHHHSSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCSSCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHSSSSSSSSCCCCCCSSSSSSSCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC MRWLWPLAVSLAVILAVGLSRVSGGAPLHLGRHRAETQEQQSRSKRGTEDEEAKGVQQYVPEEWAEYPRPIHPAGLQPTKPLVATSPNPGKDGGTPDSGQELRGNLTGAPGQRLQIQNPLYPVTESSYSAYAIMLLALVVFAVGIVGNLSVMCIVWHSYYLKSAWNSILASLALWDFLVLFFCLPIVIFNEITKQRLLGDVSCRAVPFMEVSSLGVTTFSLCALGIDRFHVATSTLPKVRPIERCQSILAKLAVIWVGSMTLAVPELLLWQLAQEPAPTMGTLDSCIMKPSASLPESLYSLVMTYQNARMWWYFGCYFCLPILFTVTCQLVTWRVRGPPGRKSECRASKHEQCESQLNSTVVGLTVVYAFCTLPENVCNIVVAYLSTELTRQTLDLLGLINQFSTFFKGAITPVLLLCICRPLGQAFLDCCCCCCCEECGGASEASAANGSDNKLKTEVSSSIYFHKPRESPPLLPLGTPC | |||||||||||||||||||||||||
| 1 | 4n6hA | 0.16 | 0.21 | 0.84 | 2.55 | Download | LEDNWETLNDNLKVIEKADN--AAQVKDALTKMRAAALDAQKATPPKLEDKSPDSPEMKDFRHGFDILVGQIDDALKLA----------NEGKVKEAQAAAEQLKTTRNAYIQKYLGSPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLM--TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALD-FRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDG-----AVVCMLQFPSP--------SWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGS--KEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG--------------------------------------------- | |||||||||||||||||||
| 2 | 4n6hA | 0.19 | 0.21 | 0.63 | 3.64 | Download | --------------------------------------------------------------------------------------------------------------------GSPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALA-TSTLPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALD-FRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRD-----GAVVCMLQFPSP--------SWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGS--KEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPC---------------------------------------------- | |||||||||||||||||||
| 3 | 4n6hA | 0.15 | 0.21 | 0.85 | 3.42 | Download | ----DLEDNWETLNDNLKVIEKADNAAQVKDALTKMRAAALDAQKATPPKLEDKSPDSPEMKDFRHGFDILVGQIDDALKLANEGKVKEAQAAAEQLKTTRNAYIQKYLGSPGA-------RSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATST-LPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFR-TPAKAKLINICIWVLASGVGVPIMVMAVTRPRD-----GAVVCMLQFP-SPS-------WYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGS--KEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG--------------------------------------------- | |||||||||||||||||||
| 4 | 2ziy | 0.12 | 0.15 | 0.74 | 1.50 | Download | -----------------------------------------------------------------------------------------------DLRDNETWWYNPSIIVHPH---WRE-FDQVPDAVYYSLGIFIGICGIIGCGGNGIVIYLFTKTKSLQTPANMFIINLAFSDFTFSLVGFPLMTISCFLKKWIFGFAACKVYGFIGGIFGFMSIMTMAMISIDRYNVIGRPMAASKKMSH-RRAFIMIIFVWLWSVLWAIGPIFGWGAYTLEG----VLCNCSFDYISRDST---------TRSNILCMFILGFFGPILIIFFCYFNIVMSVSNHEKEMAAKAQAGANAEMRLAKISIVIVSQFLLSWSPYAVVALLAQFGPLEWV---TPYAAQLPVMFAKASAIHNPMIYSVSHPKFREAISQTFPWVLCQFDDKETEDD--KDAETEIPA--GESSDAAPSADAAQMKE----- | |||||||||||||||||||
| 5 | 4n6hA | 0.16 | 0.21 | 0.84 | 3.60 | Download | ---EDNWETLNDNLKVIEKADNAAQVKDALTKMRAAALDAQKATPPKLEDKSPDSPEMKDFRHGFDILVGQIDDALKLANEGKVKEAQAAAEQLKTTRNAYIQKYLG----------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALA-TSTLPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPV-KALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRD-----GAVVCMLQFPSP--------SWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSV--RLLSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG--------------------------------------------- | |||||||||||||||||||
| 6 | 4djh | 0.19 | 0.17 | 0.60 | 1.17 | Download | -----------------------------------------------------------------------------------------------------------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVT-TTMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKAL-DFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVRE---DVDVIECSLQFPDDDYS-------WWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSGPNYREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSAA------LSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP------------------------------------------------ | |||||||||||||||||||
| 7 | 4n6hA | 0.14 | 0.21 | 0.85 | 2.86 | Download | -----DLEDNWETLNDNLKVIEKADNAAQVKDALTKMRAAALDAQKATPPKLEDKSPDSPEMKDFRHGFDILVGQIDDALKLANEG------KVKEAQAAAEQLKTTRNAYIQKYLGSPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLM--TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALD-FRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRD-----GAVVCMLQFPSP--------SWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGS--KEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG--------------------------------------------- | |||||||||||||||||||
| 8 | 4djh | 0.17 | 0.17 | 0.61 | 1.72 | Download | ---------------------------------------------------------------------------------------------------------------------------------PVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVT-TTMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLK-AKIINICIWLLSSSVGISAIVLGGTKVRE---DVDVIECSLQFPDDDYDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSGNIFEGGRITWDAYREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSA------ALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCF------------------------------------------------- | |||||||||||||||||||
| 9 | 4n6hA | 0.16 | 0.21 | 0.85 | 2.36 | Download | LEDNWETLNDNLKVIE--KADNAAQVKDALTKMRAAALDAQKATPPKLEDKSPDSPEMKDFRHGFDILVGQIDDALKLANEGKVKEAQAAAE----------QLKTTRNAYIQKYLGSPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATS-TLPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALD-FRTPAKAKLINICIWVLASGVGVPIMVMAVTRPR-----DGAVVCMLQFPSP--------SWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGS--KEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG--------------------------------------------- | |||||||||||||||||||
| 10 | 4n6hA | 0.19 | 0.17 | 0.63 | 3.54 | Download | --------------------------------------------------------------------------------------------------------------------GSPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATST-LPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPV-KALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDG-----AVVCMLQFPSP--------SWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGS--KEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG--------------------------------------------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
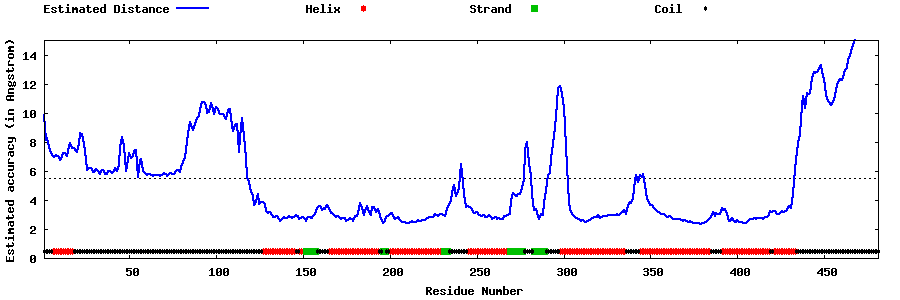
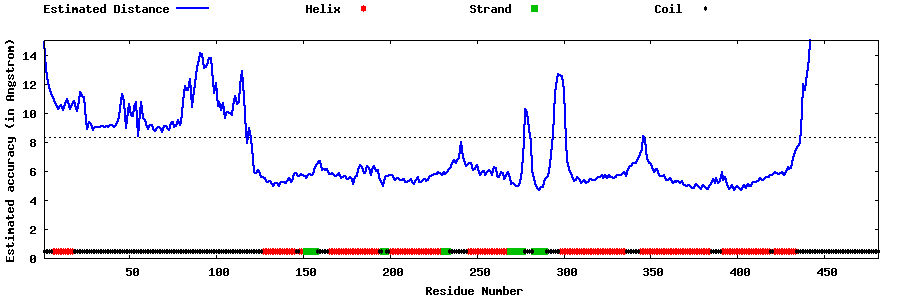
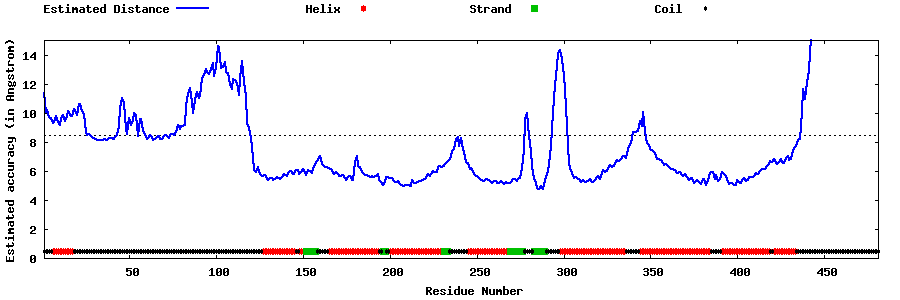
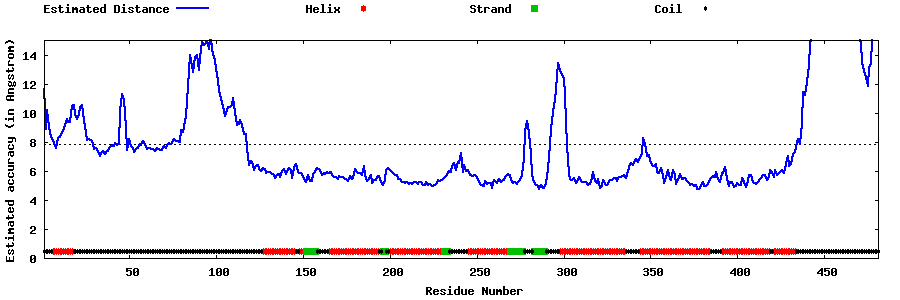
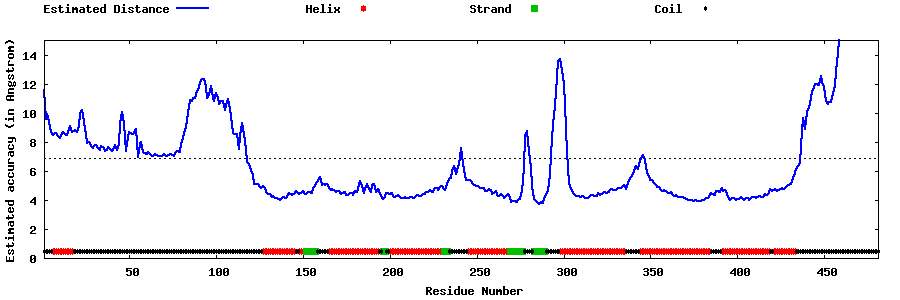
| Generated 3D models | Estimated local accuracy of models | ||
 | |||
|
|
|||
 | |||
|
| |||
 | |||
|
| |||
 | |||
|
| |||
 | |||
|
| |||