

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 | |
| | | | | | | | | | | | | | | | | | | |
| MRKMSEEEFYLFKNISSVGPWDGPQYHIAPVWAFYLQAAFMGTVFLIGFPLNAMVLVATLRYKKLRQPLNYILVNVSFGGFLLCIFSVFPVFVASCNGYFVFGRHVCALEGFLGTVAGLVTGWSLAFLAFERYIVICKPFGNFRFSSKHALTVVLATWTIGIGVSIPPFFGWSRFIPEGLQCSCGPDWYTVGTKYRSESYTWFLFIFCFIVPLSLICFSYTQLLRALKAVAAQQQESATTQKAEREVSRMVVVMVGSFCVCYVPYAAFAMYMVNNRNHGLDLRLVTIPSFFSKSACIYNPIIYCFMNKQFQACIMKMVCGKAMTDESDTCSSQKTEVSTVSSTQVGPN | |
| CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSSCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSSCCCSSSSSSSSCCCCCCCCCHHHSSSSSSHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC | |
| 998887652315777889998898657698899999999999999999987886064144068889789999999999999999997799999998198689728888999999999999999999999974202734898998388999998989999999999999998168996698489996328988641044272222429999999999999999999994413010404789999889999999999999999999999999999878752269999999999999999989999864999999999985999999987777777775332578898998 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 | |
| | | | | | | | | | | | | | | | | | | |
| MRKMSEEEFYLFKNISSVGPWDGPQYHIAPVWAFYLQAAFMGTVFLIGFPLNAMVLVATLRYKKLRQPLNYILVNVSFGGFLLCIFSVFPVFVASCNGYFVFGRHVCALEGFLGTVAGLVTGWSLAFLAFERYIVICKPFGNFRFSSKHALTVVLATWTIGIGVSIPPFFGWSRFIPEGLQCSCGPDWYTVGTKYRSESYTWFLFIFCFIVPLSLICFSYTQLLRALKAVAAQQQESATTQKAEREVSRMVVVMVGSFCVCYVPYAAFAMYMVNNRNHGLDLRLVTIPSFFSKSACIYNPIIYCFMNKQFQACIMKMVCGKAMTDESDTCSSQKTEVSTVSSTQVGPN | |
| 745344641231322143311422322112201100112311200330332020000000123301200000000103101210331131000003232000020001000100000011003000000300000001113134033310000001011101100210100001011133010200001143752311000001033113300200230332002002321553453463341212000000000100000021122000000013623010000020020000000100000000063014002410313325466644444434334344554348 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSSCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSSCCCSSSSSSSSCCCCCCCCCHHHSSSSSSHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC MRKMSEEEFYLFKNISSVGPWDGPQYHIAPVWAFYLQAAFMGTVFLIGFPLNAMVLVATLRYKKLRQPLNYILVNVSFGGFLLCIFSVFPVFVASCNGYFVFGRHVCALEGFLGTVAGLVTGWSLAFLAFERYIVICKPFGNFRFSSKHALTVVLATWTIGIGVSIPPFFGWSRFIPEGLQCSCGPDWYTVGTKYRSESYTWFLFIFCFIVPLSLICFSYTQLLRALKAVAAQQQESATTQKAEREVSRMVVVMVGSFCVCYVPYAAFAMYMVNNRNHGLDLRLVTIPSFFSKSACIYNPIIYCFMNKQFQACIMKMVCGKAMTDESDTCSSQKTEVSTVSSTQVGPN | |||||||||||||||||||||||||
| 1 | 4zwjA | 0.43 | 0.45 | 1.00 | 3.39 | Download | MCGTEGPNFYVFSNATGVSPFEYPQYYLAEPWQFSMLAAYMFLLIVLGFPINFLTLYVTVQHKKLRTPLNYILLNLAVADLFMVLGGFTSTLYTSLHGYFVFGPTGCNLQGFFATLGGEIALWSLVVLAIERYVVVCKPMSNFRFGENHAIMGVAFTWVMALACAAPPLAGWSRYIPEGLQCSCGIDYYTLKPEVNNESFVIYMFVVHFTIPMIIIFFCYGQLVFTVKEAAAQQQESATTQKAEKEVTRMVIIYVIAFLICWVPYASVAFYIFTHQGSCFGPIFMTIPAFFAKSAAIYNPVIYIMMNKQFRNCMLTTICCGKNVIFKKVSRDKGKRDYVDHVSQVEPV | |||||||||||||||||||
| 2 | 1gzmA | 0.45 | 0.43 | 0.93 | 4.41 | Download | MNGTEGPNFYPFSNKTGVSPFEAPQYYLAEPWQFSMLAAYMFLLIMLGFPINFLTLYVTVQHKKLRTPLNYILLNLAVADLFMVFGGFTTTLYTSLHGYFVFGPTGCNLEGFFATLGGEIALWSLVVLAIERYVVVCKPMSNFRFGENHAIMGVAFTWVMALACAAPPLVGWSRYIPEGMQCSCGIDYYTPHEETNNESFVIYMFVVHFIIPLIVIFFCYGQLVFTVKEAAAQQQESATTQKAEKEVTRMVIIMVIAFLICWLPYAGVAFYIFTHQGSDFGPIFMTIPAFFAKTSAVYNPVIYIMMNKQFRNCMVTTLCCGKN---DD-------------------- | |||||||||||||||||||
| 3 | 1u19A | 0.43 | 0.46 | 0.99 | 4.41 | Download | MNGTEGPNFYVPSNKTGVSPFEAPQYYLAEPWQFSMLAAYMFLLIMLGFPINFLTLYVTVQHKKLRTPLNYILLNLAVADLFMVFGGFTTTLYTSLHGYFVFGPTGCNLEGFFATLGGEIALWSLVVLAIERYVVVCKPMSNFRFGENHAIMGVAFTWVMALACAAPPLVGWSRYIPEGMQCSCGIDYYTPHEETNNESFVIYMFVVHFIIPLIVIFFCYGQLVFTVKEAAAQQQESATTQKAEKEVTRMVIIMVIAFLICWLPYAGVAFYIFTHQGSDFGPIFMTIPAFFAKTSAVYNPVIYIMMNKQFRNCMVTTLCCKNPLGDDEASTTV----SKTETSQVAPA | |||||||||||||||||||
| 4 | 2ped | 0.43 | 0.46 | 0.99 | 1.63 | Download | MNGTEGPNFYVPFSNKTGSPFEAPQYYLAEPWQFSMLAAYMFLLIMLGFPINFLTLYVTVQHKKLRTPLNYILLNLAVADLFMVFGGFTTTLYTSLHGYFVFGPTGCNLEGFFATLGGEIALWSLVVLAIERYVVVCKPMSNFRFGENHAIMGVAFTWVMALACAAPPLVGWSRYIPEGMQCSCGIDYYTPHEETNNESFVIYMFVVHFIIPLIVIFFCYGQLVFTVKEAAAQQQESATTQKAEKEVTRMVIIMVIAFLICWLPYAGVAFYIFTHQGSDFGPIFMTIPAFFAKTSAVYNPVIYIMMNKQFRNCMVTTLCCGKNPLGDDEAS---TTVSKTETSQVAPA | |||||||||||||||||||
| 5 | 2ziy | 0.22 | 0.25 | 0.99 | 1.15 | Download | -DLRDNETW--WYNPSIIVHPHWREFDQVPDAVYYSLGIFIGICGIIGCGGNGIVIYLFTKTKSLQTPANMFIINLAFSDFTFSLVGFPLMTISCFLKKWIFGFAACKVYGFIGGIFGFMSIMTMAMISIDRYNVIGRPMAASKMSHRRAFIMIIFVWLWSVLWAIGPIFGWGAYTLEGVLCNCSFDYISRD--STTRSNILCMFILGFFGPILIIFFCYFNIVMSVSNHEKEMAKAQAGANAEMRLAKISIVIVSQFLLSWSPYAVVALLAQFGPLEWVTPYAAQLPVMFAKASAIHNPMIYSVSHPKFREAISQTFPWVCQFDDKEDDKDAETEIPAGESSDAAPS | |||||||||||||||||||
| 6 | 2z73A | 0.21 | 0.24 | 0.96 | 3.34 | Download | ---------TWWYNPSIVVHPHWREFDQVPDAVYYSLGIFIGICGIIGCGGNGIVIYLFTKTKSLQTPANMFIINLAFSDFTFSLVNGPLMTISCFLKKWIFGFAACKVYGFIGGIFGFMSIMTMAMISIDRYNVIGRPMASKKMSHRRAFIMIIFVWLWSVLWAIGPIFGWGAYTLEGVLCNCSFDYIS--RDSTTRSNILCMFILGFFGPILIIFFCYFNIVMSVSNHEKEMAKAQAGANAEMRLAKISIVIVSQFLLSWSPYAVVALLAQFGPLEWVTPYAAQLPVMFAKASAIHNPMIYSVSHPKFREAISQTFPWVLTCCQFDDKETEDDKDAETEIPAGE-- | |||||||||||||||||||
| 7 | 2ped | 0.44 | 0.46 | 0.96 | 1.75 | Download | ---------VPFSNKTVRSPFEAPQYYLAEPWQFSMLAAYMFLLIMLGFPINFLTLYVTVQHKKLRTPLNYILLNLAVADLFMVFGGFTTTLYTSLHGYFVFGPTGCNLEGFFATLGGEIALWSLVVLAIERYVVVCKPMSNFRFGENHAIMGVAFTWVMALACAAPPLVGWSRYIPEGMQCSCGIDYYTPHEETNNESFVIYMFVVHFIIPLIVIFFCYGQLVFTVKEAAAQQQESATTQKAEKEVTRMVIIMVIAFLICWLPYAGVAFYIFTHQGSDFGPIFMTIPAFFAKTSAVYNPVIYIMMNKQFRNCMVTTLCCGKNPLGDDEAST---TVSKTETSQVAP- | |||||||||||||||||||
| 8 | 1u19A | 0.44 | 0.46 | 0.99 | 5.96 | Download | MNGTEGPNFYVFSNKTGVSPFEAPQYYLAEPWQFSMLAAYMFLLIMLGFPINFLTLYVTVQHKKLRTPLNYILLNLAVADLFMVFGGFTTTLYTSLHGYFVFGPTGCNLEGFFATLGGEIALWSLVVLAIERYVVVCKPMSNFRFGENHAIMGVAFTWVMALACAAPPLVGWSRYIPEGMQCSCGIDYYTPHEETNNESFVIYMFVVHFIIPLIVIFFCYGQLVFTVKEAAAQQQESATTQKAEKEVTRMVIIMVIAFLICWLPYAGVAFYIFTHQGSDFGPIFMTIPAFFAKTSAVYNPVIYIMMNKQFRNCMVTTLCCGKNPLGDDE---ASTTVSKTETSQVAPA | |||||||||||||||||||
| 9 | 2g87A | 0.44 | 0.46 | 0.99 | 3.66 | Download | MNGTEGPNFYPFSNKTGVSPFEAPQYYLAEPWQFSMLAAYMFLLIMLGFPINFLTLYVTVQHKKLRTPLNYILLNLAVADLFMVFGGFTTTLYTSLHGYFVFGPTGCNLEGFFATLGGEIALWSLVVLAIERYVVVCKPMSNFRFGENHAIMGVAFTWVMALACAAPPLVGWSRYIPEGMQCSCGIDYYTPHEETNNESFVIYMFVVHFIIPLIVIFFCYGQLVFTVKEAAAQQQESATTQKAEKEVTRMVIIMVIAFLICWLPYAGVAFYIFTHQGSDFGPIFMTIPAFFAKTSAVYNPVIYIMMNKQFRNCMVTTLCCNPLGDDEASTTVSKTETSQVAPA----- | |||||||||||||||||||
| 10 | 5dgyA | 0.43 | 0.45 | 1.00 | 4.98 | Download | MCGTEGPNFYVFSNATGVSPFEYPQYYLAEPWQFSMLAAYMFLLIVLGFPINFLTLYVTVQHKKLRTPLNYILLNLAVADLFMVLGGFTSTLYTSLHGYFVFGPTGCNLQGFFATLGGEIALWSLVVLAIERYVVVCKPMSNFRFGENHAIMGVAFTWVMALACAAPPLAGWSRYIPEGLQCSCGIDYYTLKPEVNNESFVIYMFVVHFTIPMIIIFFCYGQLVFTVKEAAAQQQESATTQKAEKEVTRMVIIYVIAFLICWVPYASVAFYIFTHQGSCFGPIFMTIPAFFAKSAAIYNPVIYIMMNKQFRNCMLTTICCGKVSRDKSVTIYLGKRDYVDHVSQVEPV | |||||||||||||||||||
| ||||||||||||||||||||||||||
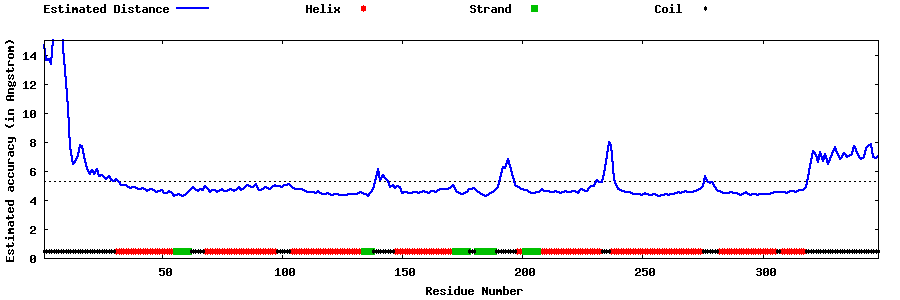
| Generated 3D models | Estimated local accuracy of models | ||
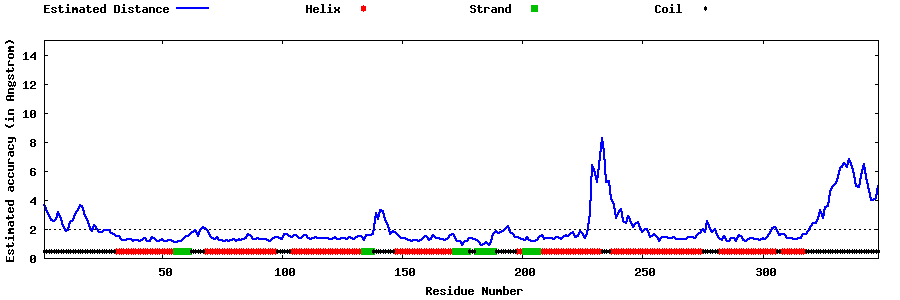 | |||
|
|
|||
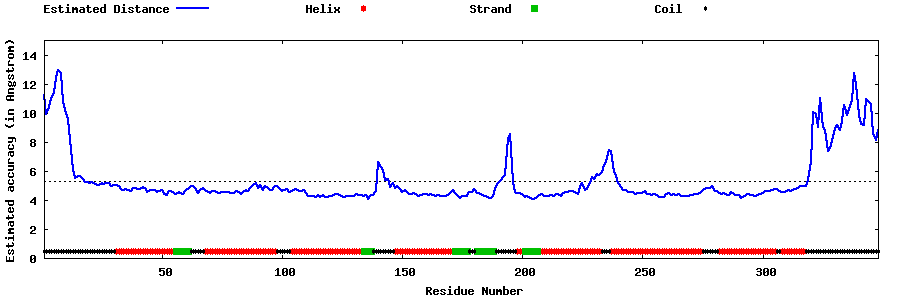 | |||
|
| |||
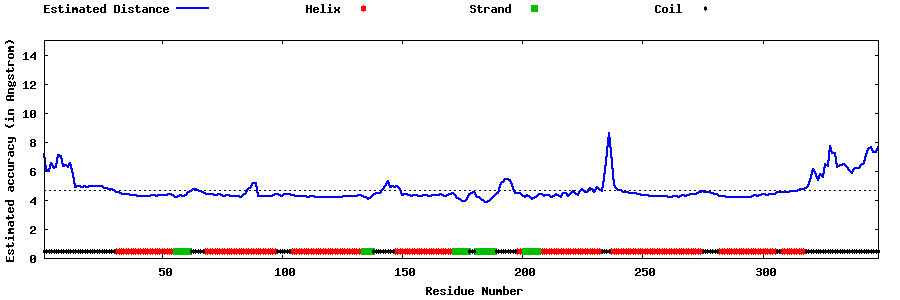 | |||
|
| |||
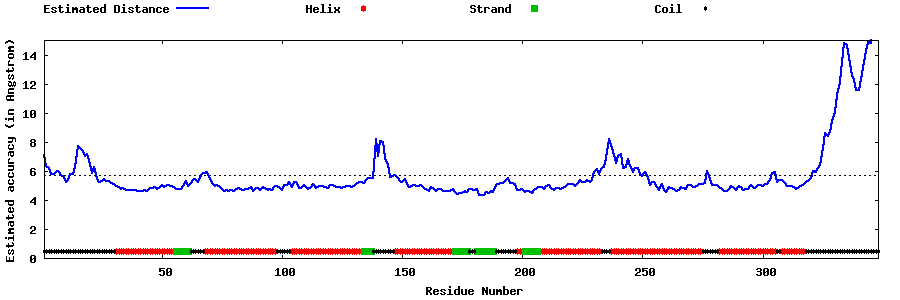 | |||
|
| |||
 | |||
|
| |||