

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | |
| | | | | | | | | | | | | | | | | | | | |
| MAQQWSLQRLAGRHPQDSYEDSTQSSIFTYTNSNSTRGPFEGPNYHIAPRWVYHLTSVWMIFVVTASVFTNGLVLAATMKFKKLRHPLNWILVNLAVADLAETVIASTISIVNQVSGYFVLGHPMCVLEGYTVSLCGITGLWSLAIISWERWLVVCKPFGNVRFDAKLAIVGIAFSWIWSAVWTAPPIFGWSRYWPHGLKTSCGPDVFSGSSYPGVQSYMIVLMVTCCIIPLAIIMLCYLQVWLAIRAVAKQQKESESTQKAEKEVTRMVVVMIFAYCVCWGPYTFFACFAAANPGYAFHPLMAALPAYFAKSATIYNPVIYVFMNRQFRNCILQLFGKKVDDGSELSSASKTEVSSVSSVSPA | |
| CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHSSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHCCSSSCCCCCSSSSSSSCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHCCCCCCCCCCCCCCCCCCSSCCCCCCCC | |
| 9731033235789997556778876522256886788988788767688899999999999999999988986264023058889799999999999999999997899999999398868608988999999999999999999999982345713888998178888760438999999999999986131563698338972236886655652688877769999999999999999999999999986277888899999999999999999999999999999999878855689999999999999999968998514899999999983888899988878887701035688999 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | |
| | | | | | | | | | | | | | | | | | | | |
| MAQQWSLQRLAGRHPQDSYEDSTQSSIFTYTNSNSTRGPFEGPNYHIAPRWVYHLTSVWMIFVVTASVFTNGLVLAATMKFKKLRHPLNWILVNLAVADLAETVIASTISIVNQVSGYFVLGHPMCVLEGYTVSLCGITGLWSLAIISWERWLVVCKPFGNVRFDAKLAIVGIAFSWIWSAVWTAPPIFGWSRYWPHGLKTSCGPDVFSGSSYPGVQSYMIVLMVTCCIIPLAIIMLCYLQVWLAIRAVAKQQKESESTQKAEKEVTRMVVVMIFAYCVCWGPYTFFACFAAANPGYAFHPLMAALPAYFAKSATIYNPVIYVFMNRQFRNCILQLFGKKVDDGSELSSASKTEVSSVSSVSPA | |
| 6445244432313334443324131321223331334212322312111101000002312210331333021000000113301200000000103000210330131000003212000031001000100000000002000000100000001113233023210000000002200310210100102012132000000001143442211000001022013300210120132002102312543454654341112000000000001110321120000000113623010000020020000000100100000043003001410423345745454444343344355458 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHSSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHCCSSSCCCCCSSSSSSSCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHCCCCCCCCCCCCCCCCCCSSCCCCCCCC MAQQWSLQRLAGRHPQDSYEDSTQSSIFTYTNSNSTRGPFEGPNYHIAPRWVYHLTSVWMIFVVTASVFTNGLVLAATMKFKKLRHPLNWILVNLAVADLAETVIASTISIVNQVSGYFVLGHPMCVLEGYTVSLCGITGLWSLAIISWERWLVVCKPFGNVRFDAKLAIVGIAFSWIWSAVWTAPPIFGWSRYWPHGLKTSCGPDVFSGSSYPGVQSYMIVLMVTCCIIPLAIIMLCYLQVWLAIRAVAKQQKESESTQKAEKEVTRMVVVMIFAYCVCWGPYTFFACFAAANPGYAFHPLMAALPAYFAKSATIYNPVIYVFMNRQFRNCILQLFGKKVDDGSELSSASKTEVSSVSSVSPA | |||||||||||||||||||||||||
| 1 | 4zwjA | 0.40 | 0.42 | 1.00 | 3.04 | Download | AKSRWYNQTTFRTGTWDAYMCGTEGPNFYVPFSNATRSPFEYPQYYLAEPWQFSMLAAYMFLLIVLGFPINFLTLYVTVQHKKLRTPLNYILLNLAVADLFMVLGGFTSTLYTSLHGYFVFGPTGCNLQGFFATLGGEIALWSLVVLAIERYVVVCKPMSNFRFGENHAIMGVAFTWVMALACAAPPLAGWSRYIPEGLQCSCGIDYYTLKPEVNNESFVIYMFVVHFTIPMIIIFFCYGQLVFTVKEAAAQQQESATTQKAEKEVTRMVIIYVIAFLICWVPYASVAFYIFTHQGSCFGPIFMTIPAFFAKSAAIYNPVIYIMMNKQFRNCMLTTICCGKNVIFKKVSRDKSVTIYLGKRDYV | |||||||||||||||||||
| 2 | 1gzmA | 0.42 | 0.38 | 0.89 | 4.14 | Download | -------------------MNGTEGPNFYVPFSNKTRSPFEAPQYYLAEPWQFSMLAAYMFLLIMLGFPINFLTLYVTVQHKKLRTPLNYILLNLAVADLFMVFGGFTTTLYTSLHGYFVFGPTGCNLEGFFATLGGEIALWSLVVLAIERYVVVCKPMSNFRFGENHAIMGVAFTWVMALACAAPPLVGWSRYIPEGMQCSCGIDYYTPHEETNNESFVIYMFVVHFIIPLIVIFFCYGQLVFTVKEAAAQQQESATTQKAEKEVTRMVIIMVIAFLICWLPYAGVAFYIFTHQGSDFGPIFMTIPAFFAKTSAVYNPVIYIMMNKQFRNCMVTTLCCGKN---DD----------------- | |||||||||||||||||||
| 3 | 2z73A | 0.23 | 0.24 | 0.92 | 4.38 | Download | ---------------------------ETWWYNPSIVVHPHWREFDQVPDAVYYSLGIFIGICGIIGCGGNGIVIYLFTKTKSLQTPANMFIINLAFSDFTFSLVNGPLMTISCFLKKWIFGFAACKVYGFIGGIFGFMSIMTMAMISIDRYNVIGRPMAASKMSHRRAFIMIIFVWLWSVLWAIGPIFGWGAYTLEGVLCNCSFDYISRDS--TTRSNILCMFILGFFGPILIIFFCYFNIVMSVSNHEKEMAAMAKGANAEMRLAKISIVIVSQFLLSWSPYAVVALLAQFGPLEWVTPYAAQLPVMFAKASAIHNPMIYSVSHPKFREAISQTFPWVLTCCQFDDKETEDDKDAETEIPAG | |||||||||||||||||||
| 4 | 2ped | 0.40 | 0.40 | 0.96 | 1.58 | Download | MNG---------TEGPNFYVPF---SNKT----GVVRSPFEAPQYYLAEPWQFSMLAAYMFLLIMLGFPINFLTLYVTVQHKKLRTPLNYILLNLAVADLFMVFGGFTTTLYTSLHGYFVFGPTGCNLEGFFATLGGEIALWSLVVLAIERYVVVCKPMSNFRFGENHAIMGVAFTWVMALACAAPPLVGWSRYIPEGMQCSCGIDYYTPHEETNNESFVIYMFVVHFIIPLIVIFFCYGQLVFTVKEAAAQQQESATTQKAEKEVTRMVIIMVIAFLICWLPYAGVAFYIFTHQGSDFGPIFMTIPAFFAKTSAVYNPVIYIMMNKQFRNCMVTTLCCGKNPLGDDEASTTVSKTETSQVAPA | |||||||||||||||||||
| 5 | 2ziy | 0.25 | 0.25 | 0.93 | 1.15 | Download | ----------------DLRDNETW------WYNPSIIVHPHWREFDQVPDAVYYSLGIFIGICGIIGCGGNGIVIYLFTKTKSLQTPANMFIINLAFSDFTFSLVGFPLMTISCFLKKWIFGFAACKVYGFIGGIFGFMSIMTMAMISIDRYNVIGRPMAASKMSHRRAFIMIIFVWLWSVLWAIGPIFGWGAYTLEGVLCNCSFDYISRDS--TTRSNILCMFILGFFGPILIIFFCYFNIVMSVSNHEKEMAKAQAGANAEMRLAKISIVIVSQFLLSWSPYAVVALLAQFGPLEWVTPYAAQLPVMFAKASAIHNPMIYSVSHPKFREAISQTFPWVQFDDKETEDDAETESSDAPSADAA | |||||||||||||||||||
| 6 | 2g87A | 0.40 | 0.40 | 0.95 | 3.09 | Download | --------------------NGTEGPNFYVPFSNVVRSPFEAPQYYLAEPWQFSMLAAYMFLLIMLGFPINFLTLYVTVQHKKLRTPLNYILLNLAVADLFMVFGGFTTTLYTSLHGYFVFGPTGCNLEGFFATLGGEIALWSLVVLAIERYVVVCKPMSNFRFGENHAIMGVAFTWVMALACAAPPLVGWSRYIPEGMQCSCGIDYYTPHEETNNESFVIYMFVVHFIIPLIVIFFCYGQLVFTVKEAAAQQQESATTQKAEKEVTRMVIIMVIAFLICWLPYAGVAFYIFTHQGSDFGPIFMTIPAFFAKTSAVYNPVIYIMMNKQFRNCMVTTLCCGKNPLGDDEASTTVSKTETSQVAPA | |||||||||||||||||||
| 7 | 2ped | 0.41 | 0.40 | 0.89 | 1.76 | Download | ----------------------------------------EAPQYYLAEPWQFSMLAAYMFLLIMLGFPINFLTLYVTVQHKKLRTPLNYILLNLAVADLFMVFGGFTTTLYTSLHGYFVFGPTGCNLEGFFATLGGEIALWSLVVLAIERYVVVCKPMSNFRFGENHAIMGVAFTWVMALACAAPPLVGWSRYIPEGMQCSCGIDYYTPHEETNNESFVIYMFVVHFIIPLIVIFFCYGQLVFTVKEAAAQQQESATTQKAEKEVTRMVIIMVIAFLICWLPYAGVAFYIFTHQGSDFGPIFMTIPAFFAKTSAVYNPVIYIMMNKQFRNCMVTTLCCGKNPLGDDEASTTVSKTETSQVAP- | |||||||||||||||||||
| 8 | 1u19A | 0.41 | 0.40 | 0.95 | 5.67 | Download | -------------------MNGTEGPNFYVPFSNKTRSPFEAPQYYLAEPWQFSMLAAYMFLLIMLGFPINFLTLYVTVQHKKLRTPLNYILLNLAVADLFMVFGGFTTTLYTSLHGYFVFGPTGCNLEGFFATLGGEIALWSLVVLAIERYVVVCKPMSNFRFGENHAIMGVAFTWVMALACAAPPLVGWSRYIPEGMQCSCGIDYYTPHEETNNESFVIYMFVVHFIIPLIVIFFCYGQLVFTVKEAAAQQQESATTQKAEKEVTRMVIIMVIAFLICWLPYAGVAFYIFTHQGSDFGPIFMTIPAFFAKTSAVYNPVIYIMMNKQFRNCMVTTLCCGKNPLGDDEASTTVSKTETSQVAPA | |||||||||||||||||||
| 9 | 2g87A | 0.41 | 0.40 | 0.95 | 3.31 | Download | -------------------MNGTEGPNFYVPFSNKTRSPFEAPQYYLAEPWQFSMLAAYMFLLIMLGFPINFLTLYVTVQHKKLRTPLNYILLNLAVADLFMVFGGFTTTLYTSLHGYFVFGPTGCNLEGFFATLGGEIALWSLVVLAIERYVVVCKPMSNFRFGENHAIMGVAFTWVMALACAAPPLVGWSRYIPEGMQCSCGIDYYTPHEETNNESFVIYMFVVHFIIPLIVIFFCYGQLVFTVKEAAAQQQESATTQKAEKEVTRMVIIMVIAFLICWLPYAGVAFYIFTHQGSDFGPIFMTIPAFFAKTSAVYNPVIYIMMNKQFRNCMVTTLCCGKNPLGDDEASTTVSKTETSQVAPA | |||||||||||||||||||
| 10 | 5w0pA | 0.40 | 0.42 | 1.00 | 4.72 | Download | QQKRWDEAAVNLAKSWDAYMCGTEGPNFYVPFSNATRSPFEYPQYYLAEPWQFSMLAAYMFLLIVLGFPINFLTLYVTVQHKKLRTPLNYILLNLAVADLFMVLGGFTSTLYTSLHGYFVFGPTGCNLQGFFATLGGEIALWSLVVLAIERYVVVCKPMSNFRFGENHAIMGVAFTWVMALACAAPPLAGWSRYIPEGLQCSCGIDYYTLKPEVNNESFVIYMFVVHFTIPMIIIFFCYGQLVFTVKEAAAQQQESATTQKAEKEVTRMVIIYVIAFLICWVPYASVAFYIFTHQGSCFGPIFMTIPAFFAKSAAIYNPVIYIMMNKQFRNCMLTTICCGDDEASAVKTETSVIFKKVSRDKSV | |||||||||||||||||||
| ||||||||||||||||||||||||||
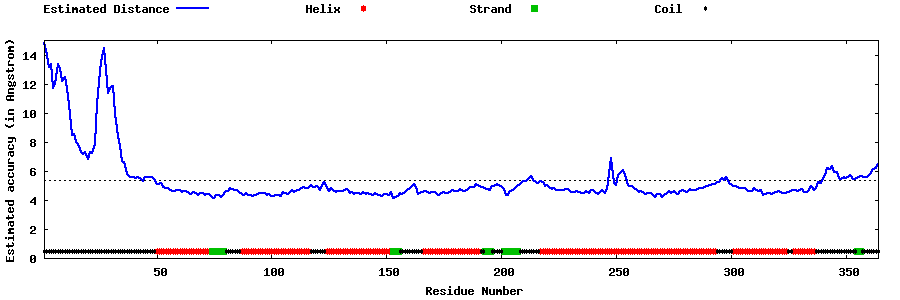
| Generated 3D models | Estimated local accuracy of models | ||
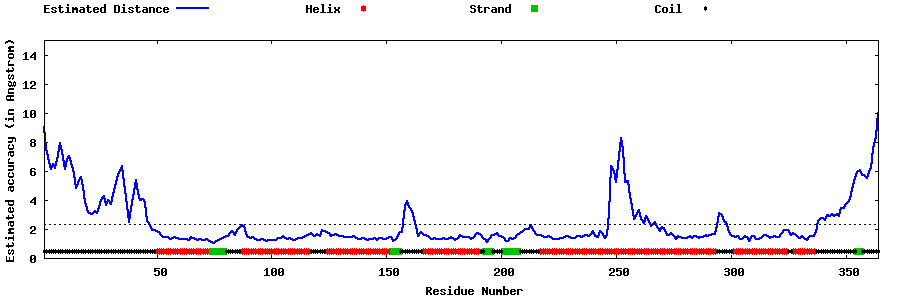 | |||
|
|
|||
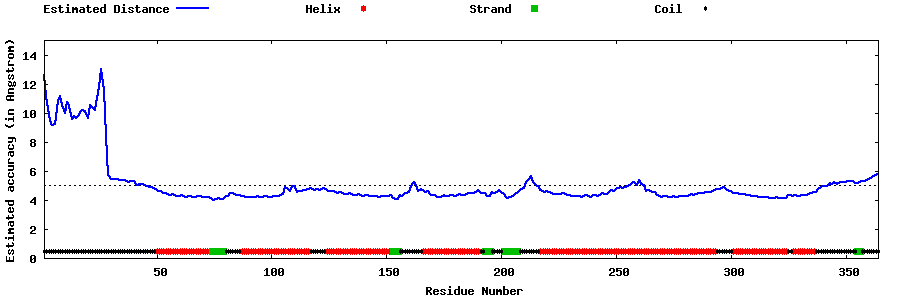 | |||
|
| |||
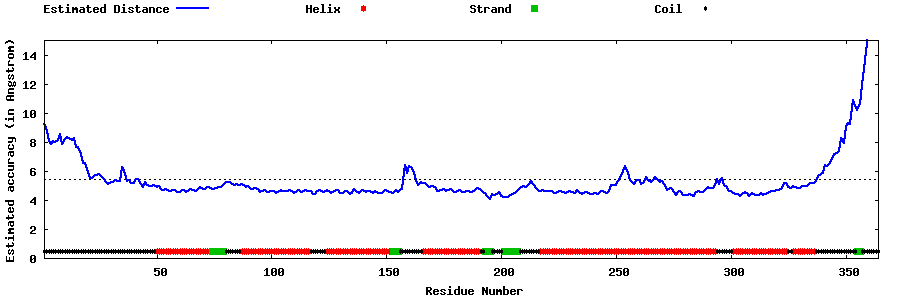 | |||
|
| |||
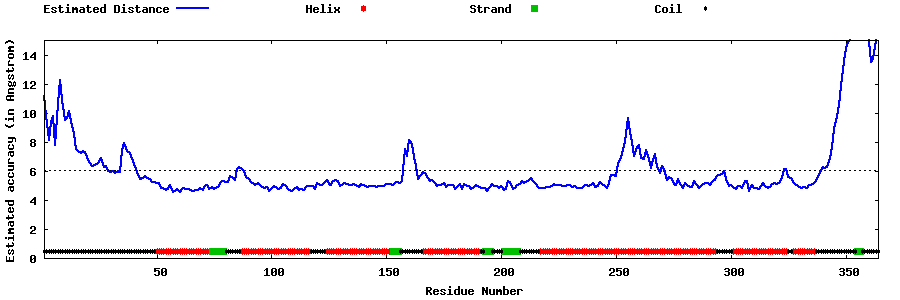 | |||
|
| |||
 | |||
|
| |||